CVR preprint reveals fundamental changes in Omicron variant
Published: 5 January 2022
A Centre for Virus Research study of the Omicron variant of SARS-CoV-2 has shown that it represents a step-change in the COVID-19 pandemic, revealing the different route it uses to infect human cells.
A Centre of Virus Research study of the Omicron variant of SARS-CoV-2 has shown that it represents a step-change in the COVID-19 pandemic, revealing the different route it uses to infect human cells.
The work, led by Professor Emma Thomson and published as a pre-print study, has used laboratory experiments and real world infection data to investigate Omicron.
Record numbers of new daily infections are being reported across the globe - but quite why Omicron is spreading so rapidly remains unknown.
However, this CVR research has revealed fundamental changes that are likely to be important for Omicron spread and disease.
Laboratory tests found that Omicron is largely unaffected by the antibodies provided by two doses of COVID-19 vaccine, but responses are improved by a third dose booster. Importantly, this is confirmed by analysis of real world infection data; Omicron escape immunity from two vaccine doses, but three doses restores good levels of protection.
The work also provides evidence that Omicron has switched its route of entry into human cells, from cell surface fusion to cathepsin-dependent fusion within the endosome. This fundamental biological shift is likely to influence Omicron spread and the types of cells it can hijack.
These changes may also affect the pathogenesis and severity of disease, and researchers say, require further evaluation in population-based studies.
Professor Thomson said: "In our paper, we describe a fundamental shift in antigenicity and cell entry of the SARS-CoV-2 Omicron variant. "A series of experiments carried out by Joe Grove and others at the CVR showed that Omicron enters cells independently of TMPRSS2 via the endosomal pathway.
"That is a huge phenotypic change and might affect cell tropism and clinical severity, with more data to follow on this soon.
"Part of the work led by Professor Brian Willet showed marked drops in neutralisation of Omicron in recipients of two doses of deployed vaccines in the DOVE study.
"A third dose of deployed vaccine - a booster dose of BNT162b2 or half-dose mRNA-1273 - improved these responses. The take-home message here is that booster doses are really important.
"The changes in effectiveness of two doses of vaccine means that Omicron is likely to transmit more easily and boosting is really important. Three vaccine doses makes a huge difference to immunity, resulting in the same take-home message - boosters are really important for immunity to Omicron.”
"While Omicron may cause slightly less severe illness, deaths and hospitalisations are not fully decoupled from infection and hospitals are under rising pressures.”
"This was a huge team effort, and very many thanks go to the scientists, clinicians, stats team & bioinformaticians from the CVR, University of Glasgow School of Mathematics & Statistics, NHS Greater Glasgow and Clyde, NHS Lothian and others, all of whom worked extremely hard over the holiday period.
"A huge thank you also to our participants for giving up their time to take part our studies and to the incredible Glasgow Research Facility nurses, doctors, admin team and managers for going above and beyond to get these studies done."
'The hyper-transmissible SARS-CoV-2 Omicron variant exhibits significant antigenic change, vaccine escape and a switch in cell entry mechanism'
- Brian J. Willett, Joe Grove, Oscar A MacLean, Craig Wilkie, Nicola Logan, Giuditta De Lorenzo, Wilhelm Furnon, Sam Scott, Maria Manali, Agnieszka Szemiel, Shirin Ashraf, Elen Vink, William Harvey, Chris Davis, Richard Orton, Joseph Hughes, Poppy Holland, Vanessa Silva, David Pascall, Kathryn Puxty, Ana da Silva Filipe, Gonzalo Yebra, Sharif Shaaban, Matthew T. G. Holden, Rute Maria Pinto, Rory Gunson, Kate Templeton, Pablo Murcia, Arvind H. Patel, John Haughney, David L. Robertson, Massimo Palmarini, Surajit Ray, & Emma C. Thomson
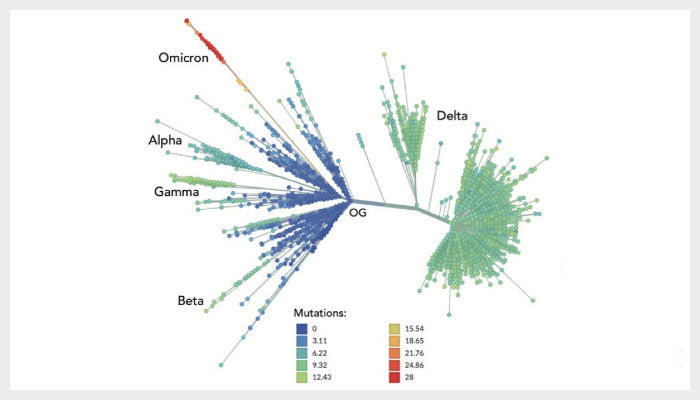
Image legend: Figure 1 - Spike amino acid changes typifying the Omicron variant. (C) Inferred evolutionary relationships of SARS-CoV-2 from NextStrain with the Variants of Concern labelled. The colours of the tree tips correspond to the number of mutations causing Spike amino acid substitutions relative to the SARS-CoV-2 original genotype (OG) reference strain Wuhan-Hu-1.
Enquiries: ali.howard@glasgow.ac.uk or elizabeth.mcmeekin@glasgow.ac.uk / 0141 330 6557 or 0141 330 4831
First published: 5 January 2022

