Natural origins of SARS-CoV-2
Published: 9 February 2022
New research by Centre for Virus Research (CVR) scientists has disentangled the complex evolutionary history of the bat coronaviruses related to SARS-CoV-2, the virus responsible for COVID-19.
New research by Centre for Virus Research (CVR) scientists has disentangled the complex evolutionary history of the bat coronaviruses related to SARS-CoV-2, the virus responsible for COVID-19.
This work, published in Genome Biology and Evolution, demonstrates that accounting for recombination (viruses mixing their genetic material) in the past evolutionary history of these SARS-related coronaviruses recombinants is important for reliably inferring relatedness to SARS-CoV-2.
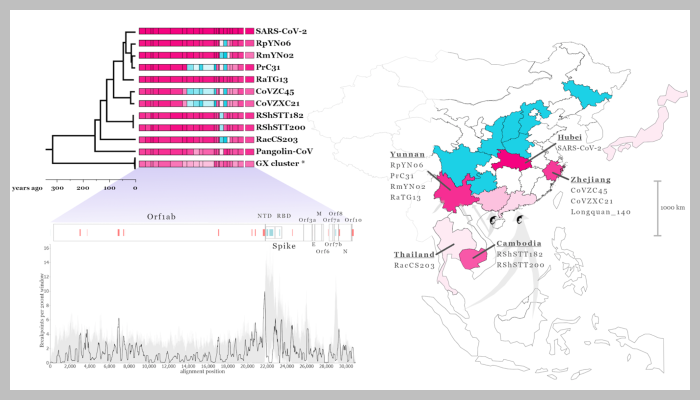
This is exemplified by the RaTG13 bat virus, which is often described as the on average closest virus to SARS-CoV-2, despite by far not being the evolutionary closest virus once recombination history is accounted for in the other SARS-related coronaviruses. This means that the proximal ancestor of SARS-CoV-2 could be distributed over a large geographic area and is not necessarily from Yunnan as is often - incorrectly - assumed.
While scientists agree that bats are the most likely animal host species of SARS-CoV-2 ancestors, no one has yet been able to fully explain the virus’s history or pinpoint the animal species that ultimately passed the new coronavirus to humans.
Now, in this research led by the CVR in collaboration with a team of international scientists, researchers have been able to find out more than ever before about the evolutionary history of SARS-CoV-2, unambiguously tracing the virus origins to horseshoe bats. The new study is published in Genome Biology and Evolution.
This latest research takes a look at the viral evolution of the SARS-related coronaviruses by investigating the history of co-circulating bat coronaviruses and the phenomenon of virus recombination – when a host bat is infected with two coronaviruses at once and the viruses swap bits of their genetic material inside host cells to create a new, so-called recombinant virus (a similar outcome to sex in higher organisms).
Recombination is common, and the process is used by many viruses, including coronaviruses, to generate novel genome forms contributing to adaptation and persistence.
Spyros Lytras explains abstract for Exploring the natural origins of SARS-CoV-2
CVR PhD student Spyros Lytras, first author of the study, said: “Recombination is a mechanism seen in many viruses, where related viruses can swap bits of their genetic material when they find themselves infecting the same cell, resulting in a new mosaic virus.
"It is clear that recombination is a defining feature of SARS-related coronaviruses and essential to account for when examining the viruses’ evolution.”
SARS-related coronaviruses, like SARS-CoV-2, are dispersed over a large geographical area across China and Southeast Asia. Since the 2020 coronavirus pandemic began, scientists have uncovered a number of viruses related to SARS-CoV-2, retrieved from locations in China and Southeast Asia. So far, some of the closest known virus relatives to SARS-CoV-2 were collected in Yunnan, China, and Laos, but they only shared a common ancestor with the newest coronavirus a little less than 40 years ago.
However, several of the viruses related to SARS-CoV-2 are known to be recombinant viruses, suggesting the virus responsible for COVID-19 does not have one single and simple evolutionary line.
Professor David L Robertson, senior author on the study, said: “Worryingly this indicates there is undoubtedly a virus highly related to SARS-CoV-2 still present somewhere in the wild in China.”
This research analysed a wide distribution of related viruses in bats, with shared markers of viral recombination (specific points in the virus’s genetic material, or RNA) suggesting that SARS-CoV-2 ancestors were frequently transmitting amongst bats species, with the virus changing and recombining over time as host bats were infected with two coronaviruses at once.
The study also shows that there were multiple recombination events in SARS-CoV-2’s past, taking place in the past few centuries across an extensive geographic area of at least 2,500km, and the researchers agree that more work needs to be done to pinpoint closer relatives of SARS-CoV-2 by increasing virus sampling in wildlife.
Professor Robertson added: “Our analysis highlights the need for dramatically more wildlife sampling to pinpoint the exact origins of SARS-CoV-2 and understood more fully the risk of infection of humans by viruses like these across China and Southeast Asia.
"The finding of bat coronaviruses that can so readily use the human - and other mammals - ACE2 receptor without having to undergo any significant evolutionary change underscores the inevitability of future spillovers.
"More wildlife sampling would also help identify the extent of the diversity in the related coronaviruses circulating in the wild that may present high risk for future spill overs.”
Exploring the natural origins of SARS-CoV-2 in the light of recombination
- Spyros Lytras, Joseph Hughes, Darren Martin, Phillip Swanepoel, Arné de Klerk, Rentia Lourens, Sergei L Kosakovsky Pond, Wei Xia, Xiaowei Jiang, David L Robertson
- Genome Biology and Evolution, evac018, https://doi.org/10.1093/gbe/evac018 Published: 08 February 2022.
Figure legend: Recombination analysis of the bat and pangolin SARS-related coronaviruses genomes most closely related to SARS-CoV-2 where pink indicates proximity to SARS-CoV-2 and blue proximity to SARS-CoV. The phylogenetic tree is from the fifth recombination-minimised region, and the scale below shows time to most recent common ancestor. Below this genome regions are shown above a plot of the distribution of recombination breakpoints; hotspots are indicated by red and coldspots by blue lines. The map to the right shows the sampling locations of these SARS-related coronaviruses; shading intensity indicates relative proximity to SARS-CoV-2.
Funding: The work was funded by the Medical Research Council (MRC) and the Wellcome Trust.
Contacts: For more information, contact Elizabeth McMeekin or Ali Howard in the University of Glasgow Communications and Public Affairs Office on 0141 330 4831 or 0141 330 6557; or email Elizabeth.mcmeekin@glasgow.ac.uk or ali.howard@glasgow.ac.uk
First published: 9 February 2022

