Machine learning can predict animal viruses that risk infecting humans
Published: 4 October 2021
Centre for Virus Research scientists have developed a new machine learning method that can accurately predict which animal viruses could go on to infect humans in the future, using only information encoded in the viral genome.
Centre for Virus Research scientists have developed a new machine learning method that can accurately predict which animal viruses could go on to infect humans in the future, using only information encoded in the viral genome.
Most emerging infectious diseases of humans are caused by ‘zoonotic’ viruses that originate from other animal species.
Of the many millions of viruses that circulate in animals, however, only a few are likely able to infect humans.
Scientists currently have very limited ability to rapidly assess zoonotic risk at the time that viruses are discovered, making it difficult to know which newly discovered viruses should be prioritised for early investigation, and beyond that, outbreak preparedness.
Now, in a new study led by the CVR and published in PLOS Biology, researchers have developed a new machine learning method for accurately predicting which viruses could infect humans, based entirely on a virus’s genome sequence – often the only thing scientifically known about newly found or poorly-characterised animal viruses - as well as ranking them as 'low', 'medium', 'high', or 'very high' risk.
Using the same modelling, and without any prior knowledge of the previous SARS outbreak in humans, this model was also able to accurately predict that SARS-CoV-2, the virus that caused the COVID-19 pandemic, and its closest viral relatives found in animals, had a high risk of being able to infect humans.
This finding, together with more formal testing on hundreds of viruses with known zoonotic status, showed that the model makes actionable predictions on a diverse range of RNA and DNA viruses, even those that are entirely new to science.
The new modelling method predicts whether viruses might be able infect humans, but cannot determine how dangerous they may be in terms of either symptoms or epidemic/pandemic potential, nor when they might jump into human populations.
Being able to infect humans is the first step towards causing an outbreak, but numerous other factors, such as contact between the reservoir and humans, whether the virus can transmit between humans, and our response to such ‘spillover infections’ will shape epidemic and pandemic risk.
Researchers believe this new modelling method could help scientists better prioritise research efforts on the animal viruses most likely to successfully infect humans, an important step towards future human outbreak preparedness and planning.
Lead author Dr Nardus Mollentze, Research Associate at the CVR, said: “Calls for investment in virus discovery programmes targeting wildlife have been controversial, since it remains unclear how to go from knowing which viruses are out there to outbreak preparedness.
"Finding out what newly described viruses are capable of, and how to respond to that, requires extensive characterisation in both the lab and in their natural environment, and this characterisation currently cannot keep up with the number of viruses being found.
"When viruses are first discovered, often all we have is their genome sequence, so developing an accurate machine learning tool that is based on information contained within that should enable us to better understand which animal viruses pose the highest risk, and should therefore be characterised and investigated first.
“Such predictions are still only a first step however. If we want investments in virus discovery to translate into pandemic preparedness, there is a need to develop both higher-throughput virus characterisation methods and further models capable of turning the information generated by these methods into updated risk predictions.”
Senior author Dr Daniel Streicker, from the CVR and the Institute of Biodiversity, Animal Health and Comparative Medicine, said: “Identifying high risk viruses amid the vast diversity of animal-infecting viruses that are unlikely to infect humans has been a needle in a haystack challenge. Our new genome-based zoonotic risk assessment represents a step towards solving that challenge and, along with our earlier efforts showing that the reservoir hosts and arthropod vectors of viruses can be predicted from viral genomes, shows that a surprising amount of ecological insight is possible from genome sequences alone, hinting at the existence of poorly understood ways that viruses adapt to their hosts.
“More immediately, since these models use nothing more than genetic sequences, they can be applied at the time that viruses are discovered, creating a rapid, low-cost triage system to decide which viruses merit extra attention.”
Co-author Dr Simon Babayan, Institute of Biodiversity, Animal Health and Comparative Medicine, said: “As most emerging infectious diseases in humans are caused by a small number of viruses that originated in other animal species, it remains an enormous challenge to know where to look for the next virus epidemic.
"Now we provide a rapid, low-cost approach to enable evidence-driven virus surveillance and characterisation of viruses that could specifically infect humans, and may, therefore, better help with future epidemic and pandemic preparedness.”
Identifying and prioritizing potential human-infecting viruses from their genome sequences
- Nardus Mollentze, Simon A. Babayan, Daniel G. Streicker
- Published: September 28, 2021. https://doi.org/10.1371/journal.pbio.3001390
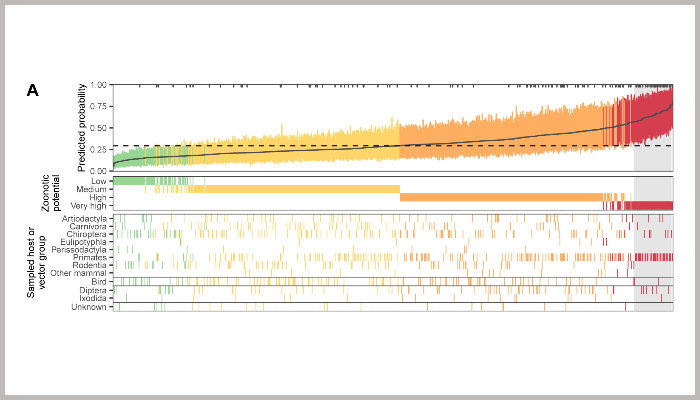
Image legend: Figure 3. Probability of human infection predicted from holdout viral genomes.
(A) Predicted probability of human infection for 758 virus species that were not in the training data. Colors show the assigned zoonotic potential categories, with an additional panel showing the host or vector group each virus genome was sampled from. Tick marks along the top edge of the first panel show the location of virus genomes sampled from humans, while a dashed line shows the cutoff that balanced sensitivity and specificity in the training data.
Funding: The work was funded by the Medical Research Council (MRC) and Wellcome.
Enquiries: ali.howard@glasgow.ac.uk or elizabeth.mcmeekin@glasgow.ac.uk / 0141 330 6557 or 0141 330 4831
First published: 4 October 2021

