Dr Christopher Quince
My research spans both bioinformatics and mathematical modelling applied to microbial communities. Environmental genomics coupled to next-generation sequencing has transformed microbial ecology. Direct sequencing of DNA can reveal both community composition but also function. Sequencing of the 16S rRNA gene following PCR amplification has revealed striking levels of diversity in many environments. However, it is vital to distinguish noise from true variation when determining diversity. I have developed a software pipeline, AmpliconNoise, for removing noise from 454 pyrosequenced amplicons. I am now extending these algorithms to other sequencing platforms principally MiSeq and IonTorrent. Shotgun metagenome data, is potentially much more informative than 16S rRNA, as functional genes can be obtained but this added complexity makes interpretation difficult. As part of a TSB funded project in collaboration with Unilever I am developing a robust easy-to-use software pipeline for processing of shotgun metagenome data specifically for commercial applications. My long-term goal is to link metagenomics with others forms of high-throughput data, e.g. metatranscriptomics and metabolomics, in order to parameterise mathematical models capable of predicting community function. I apply these methods to a wide-range of systems from soil to the human microbiome. I am the recipient of a prestigious EPSRC Career Acceleration Fellowship to apply genomics to environmental engineering systems. We have succeeded in characterising the microbial diversity of water treatment filters and are now using that information to manipulate communities for improved pollutant removal.
Current project:
Modelling Synthetic Microbial Communities.
For synthetic biology to reach its full potential, experimental advances must be accompanied by the development of mathematical tools capable of predicting the functioning of the designed organisms. This will enable organisms to be optimised for a specific role e.g. to maximise production of a desired compound. To model an organism’s metabolism, gene expression and regulation completely is extremely challenging. It may be possible in some very well studied systems, e.g. E. coli, but when that information is absent, flux balance analysis (FBA), which requires only the topology of the metabolic network, has proved useful for identifying potential bottle-necks that can be targeted to increase production. Many processes in environmental engineering, for example anaerobic digestion (AD), the conversion of organic waste into biogas, a renewable form of energy, involve not a single organism but a community of interacting organisms. However, the extension of these models, to multiple species, some potentially naturally occurring and others designed presents even more challenges. It is then important to consider how the different populations will interact with each other. We are developing models capable of predicting the function and dynamics of these communities from a combination of metabolic networks for the individual species and kinetic parameterisation from growth experiments. The approach is to couple the metabolic models through substrates and products in the environment with demographic processes controlling the growth of populations (Figure 1). We are validating the models through simple experiments on synthetic communities of known organisms capable of performing AD. Once developed this will provide a framework for optimising synthetic AD communities for biogas production.
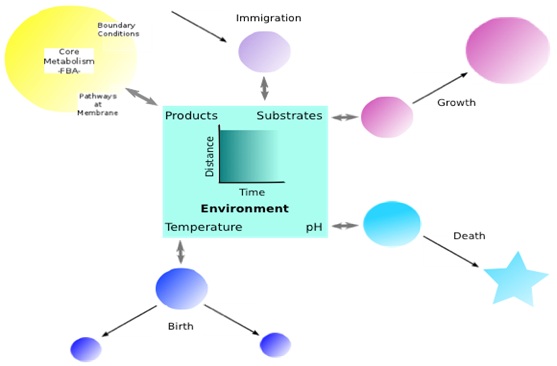
Fig. 1 schematic of model structure linking individuals from different species with different metabolisms to the environment
Academic and industrial links:
I have extensive links with Unilever, two students and one PDRA, are funded by them through a direct grant my group to develop bioinformatics tools for microbial community analysis. As mentioned above we also collaborate with Unilever through a joint TSB grant. I have links with the water industry both in the UK and India. I have collaborations with Scottish and Northumbrian Water studying microbial communities in water treatment plants and a collaboration with pharmaceutical manufactures in Mumbai to develop better water treatment methods for pharmaceutical waste. My academic links span both national and international universities. I collaborate with a mathematician Dr Todd Parsons at Marie-Curie Universitie in Paris. I work with numerous groups in the United States, for example Dr Peter Turnbaugh in Harvard. I am jointly supervising students both in Stockholm (SciLifeLab) and in Australia (Hawkesbury Institute for the Environment). I work with U.K. based researchers at Liverpool, Newcastle, Bangor and Aberdeen and of course the University of Glasgow itself where I work with researchers from engineering, biology and medicine.

