CVR scientists characterise first significant SARS-CoV-2 mutation
Published: 31 January 2021
An international collaboration involving Centre for Virus Research scientists Professor David Robertson, Professor Emma Thomson and Dr Ana Filipe has characterised the first significant SARS-CoV-2 mutation, N439K, occurring within the virus’s spike protein receptor binding motif (RBM).
An international collaboration involving Centre for Virus Research (CVR) scientists Professor David Robertson, Professor Emma Thomson and Dr Ana Filipe has characterised the first significant SARS-CoV-2 mutation, N439K, occurring within the virus’s spike protein receptor binding motif (RBM).
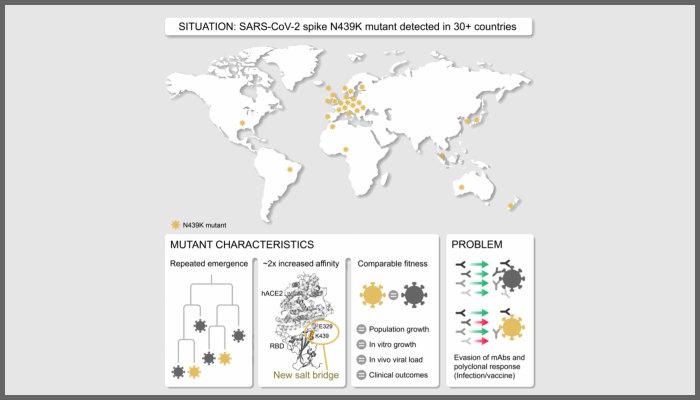
The N439K mutation, originally discovered in the first half of 2020, is considered to be one of the first important SARS-CoV-2 receptor binding motif (RBM) mutations, because it occurred within the receptor binding domain (RBD) in the virus’s Spike protein, which interacts with the human ACE2 receptor.
This study, published in the journal Cell, shows how this change in SARS-CoV-2’s genome increases the binding affinity of the virus to the human ACE2 receptor.
The frequent occurrence of mutations within the RBM highlights the ‘plasticity’ of this part of the Spike protein, which is a major target of vaccines and therapeutic monoclonal antibodies.
Researchers have characterised the effect of the N439K mutation from a combined molecular and protein structure perspective, placing this knowledge into a detailed epidemiological, clinical and evolutionary context.
When the study began in June 2020, the relatively slow rate of evolution was giving scientists the impression that SARS-CoV-2 mutations would result in no immediate threat to vaccines or therapies.
Senior author Professor David Robertson, Head of Viral Genomics and Bioinformatics at the CVR, said: “The amino acid change N439K was the first RBM amino acid replacement that increased to high frequency and so can be viewed as a ‘sentinel’ mutation for SARS-CoV-2 antigenic change.
"By studying it in detail, we intended to better understand the potential evolutionary trajectory of the new virus.”
This first lineage defined by N439K was, other than a small number of cases, only observed in Scotland. As a result of the lockdown in the UK, this lineage - along with many others - was driven to extinction, demonstrating that lockdowns not only decrease infection numbers but can impact on virus variation too.
At the time, one other virus had been observed with the N439K mutation. Unfortunately, these variants have now seeded the global spread of variants carrying this RBM change to more than 30 countries.
Co-author Professor Emma Thomson said: “This spread is unlikely to be anything to do with the N439K mutation as we observe no increased - or decreased - fitness relative to wildtype.”
Out of concern for the potential harm N439K variants may cause, Professor Thomson activated a clinical team to look into the clinical history of infected Scottish individuals.
The Professor in Infectious Diseases added: “As members of the COG-UK consortium, and due to the hard work of our genome sequencing team led by Dr Ana Filipe, we had a large number of SARS-CoV-2 genomes available so could identify cases associated with the mutated virus and compare their hospital presentation.
"Reassuringly, no increased disease severity was observed but at the same, the N439K variants seemed to be just as virulent as wildtype in patients.”
This ‘successful’ nature of the variant was subsequently confirmed by experimental work at the CVR. Although N439K variants were not found to be more virulent or more transmissible than the original SARS-CoV-2 strain, this research was the first to demonstrate that mutations that maintain viral fitness can evade immunity.
Professor Thomson explained: “We were concerned about this mutation in 2020 because it was predicted to increase binding to the human ACE2 receptor and we were worried that it might be associated with escape from natural immunity.
"So we were initially relieved when it disappeared after the first lockdown. However, it became clear the virus could mutate to avoid the immune response without losing fitness to replicate.
"We were able to demonstrate this in our high biocontainment laboratories through cross-competition assays at the CVR.”
To understand if and how the N439K mutation might evade immunity, researchers at the CVR collaborated with researchers at Vir Biotechnology, based in Switzerland and the USA, who quantified the plasticity of SARS-CoV-2 RBM region and conducted detailed protein structural analysis, including solving the bound N439K-mutation-human-ACE2 structure.
Senior author Gyorgy Snell, of Vir Biotechnology, said: “Looking at the how Spike’s binding interface made contact with the ACE2 receptor, it was immediately clear SARS-CoV-2 was going to accommodate further amino acid changes contributing to immune evasion and more significant changes than N439K was causing.”
Professor Robertson added: “Our results here on the plasticity of the RBM predicted the emergence of many of the changes present in the SARS-CoV-2 variants of concern.
"Collectively, these data indicate the importance of rolling out the vaccine to as many people as is possible as quickly as possible in the coming months and of genome surveillance efforts to monitor for novel variants with new antigenic properties.
"There is no evidence to indicate that the vaccine won’t remain effective in the short to medium term but what is happening is the circulating SARS-CoV-2 are moving away from the early variants that were used in the first vaccine preparations.”
Professor Thomson said: “The plasticity of the receptor binding motif in the spike protein of SARS-CoV-2 and the evidence that a single mutation, N439K, can impair neutralizing antibody responses indicates that we may need to update vaccine design in the future, particularly in the light of the emergence of new variants around the world.
"The importance of our early-warning sequencing surveillance system, carried out across the UK by the COG-UK consortium, is very clear.
"Understanding the impact of new variant strains that have multiple changes in the genome on vaccines is the next major challenge for the scientific community.
Professor Robertson concluded: “The need for vaccine re-design is looking inevitable, particularly as the vaccine coverage is going to be very unevenly distributed.
"We urgently need a more equitable use of vaccines internationally with scale up in production and dissemination. Controlling this virus is possible - but it needs a more coordinated global effort.”
Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity
- Emma C. Thomson, Laura E. Rosen, James G. Shepherd, Roberto Spreafico, Ana da Silva Filipe, Jason A. Wojcechowskyj, Chris Davis, Luca Piccoli, David J. Pascall, Josh Dillen, Spyros Lytras, Nadine Czudnochowski, Rajiv Shah, Marcel Meury, Natasha Jesudason,, Anna De Marco, Kathy Li, Jessica Bassi, Aine O’Toole, Dora Pinto, Rachel M. Colquhoun, Katja Culap, Ben Jackson, Fabrizia Zatta, Andrew Rambaut, Stefano Jaconi, Vattipally B. Sreenu, Jay Nix, Ivy Zhang, Ruth F. Jarrett, William G. Glass, Martina Beltramello, Kyriaki Nomikou, Matteo Pizzuto, Lily Tong, Elisabetta Cameroni, Tristan I. Croll, Natasha Johnson, Julia Di Iulio, Arthur Wickenhagen, Alessandro Ceschi, Aoife M. Harbison, Daniel Mair, Paolo Ferrari, Katherine Smollett, Federica Sallusto, Stephen Carmichael, Christian Garzoni, Jenna Nichols, Massimo Galli, Joseph Hughes, Agostino Riva, Antonia Ho, Marco Schiuma, Malcolm G. Semple, Peter J.M. Openshaw, Elisa Fadda, J. Kenneth Baillie, John D. Chodera, Suzannah J. Rihn, Samantha J. Lycett, Herbert W. Virgin, Amalio Telenti, Davide Corti, David L. Robertson, Gyorgy Snell.
- DOI:https://doi.org/10.1016/j.cell.2021.01.037.
Enquiries: ali.howard@glasgow.ac.uk or elizabeth.mcmeekin@glasgow.ac.uk / 0141 330 6557 or 0141 330 4831
First published: 31 January 2021

