NMR spectroscopy
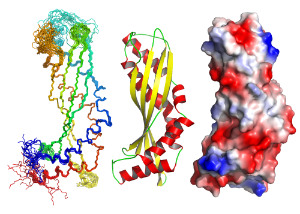
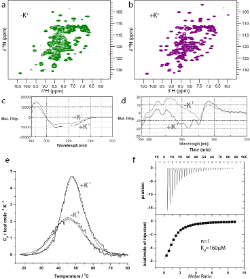
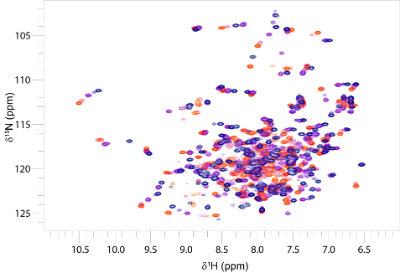 Biomolecular nuclear magnetic resonance spectroscopy (NMR) is a very versatile technique that can be used to determine the three dimensional structures of proteins, DNA, RNA, oligosaccharides and small molecules. It can also be used to study conformational changes, intermolecular interactions (both strong and weak), and molecular motion on many timescales in solution. NMR is widely used in metabolomics studies. (and enquiries for this type of application should be directed to Glasgow Polyomics) Solution state NMR complements the other techniques, such as circular dichroism and X-ray crystallography, available in the facility.
Biomolecular nuclear magnetic resonance spectroscopy (NMR) is a very versatile technique that can be used to determine the three dimensional structures of proteins, DNA, RNA, oligosaccharides and small molecules. It can also be used to study conformational changes, intermolecular interactions (both strong and weak), and molecular motion on many timescales in solution. NMR is widely used in metabolomics studies. (and enquiries for this type of application should be directed to Glasgow Polyomics) Solution state NMR complements the other techniques, such as circular dichroism and X-ray crystallography, available in the facility.
Generally, biomacromolecular NMR samples should be prepared with highly purified material, often isotopically labelled with 15N, 13C and even 2H, but NMR has been used e.g. to study proteins in living cells, and ligand-receptor interactions where the receptor comprises an un-purified membrane preparation.
Some guidance on typical sample requirements are given below, but given the breadth of applications possible with NMR, and the variety of specific sample requirements, it is advised that you discuss your project with Dr Brian Smith before planning experiments.
 |
 |
Typical Sample Requirements
For biomacromolecules (proteins, DNA and RNA), samples should typically be >95% pure in low (100 mM) ionic strength, aqueous buffer that contains only hydrogen atoms that do exchange with the solvent, e.g. sodium phosphate.
The sample should be stable in solution for hours to weeks at the required temperature (typically 15-40 °C)
The buffer used should not contain additives such as glycerol, detergents and reducing agents. Alternatively, deuterated buffer components (e.g. Tris, HEPES, imidazole, EDTA, DTT, TCEP) should be used.
Biomacromolecule concentrations should ideally be in the range 0.1-2.0 mM. The standard NMR sample volume is 600 ml meaning that 60 nmoles to 1.2 mmoles of the target will be required per sample.

