Systematic Reconstruction of Complete TC Sensorial Network in Staphylococcus aureus
Published: 17 September 2020
A project developed by Institute researcher Dr Andreas Haag has produced the first ever complete reconstruction of the two-component signal transduction network of the major human pathogen Staphylococcus aureus.
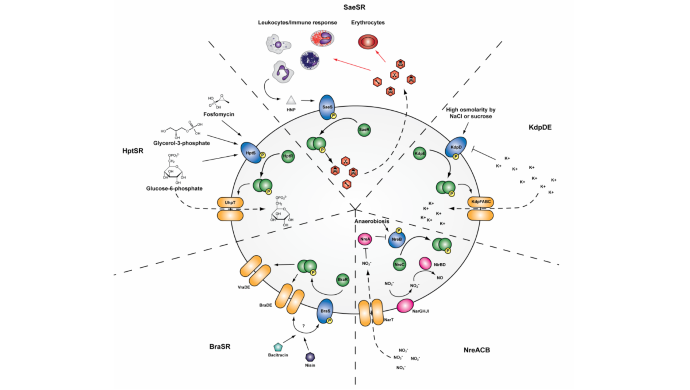
A project developed by Institute researcher Dr Andreas Haag has produced the first ever complete reconstruction of the two-component signal transduction network of the major human pathogen Staphylococcus aureus.
The research, published in mSystems, is the product of a collaboration with the lab of Professor Inigo Lasa at the University of Navarra, Spain and Dr Virginia De Cesare at the MRC PPU at the University of Dundee.
The data and tools developed provide valuable insights into the regulatory mechanisms of S. aureus and will continue to help in the understanding of how the pathogen evades host immunity.
Identifying how bacteria, and in particular pathogenic bacteria, sense and respond to their environment is key to understanding how they survive fluctuating or adverse conditions and cause disease.
Bacteria can sense environmental conditions and respond accordingly; the sensorial system responsible relies on pairs of sensory and regulatory proteins, known as two-component systems (TCSs).
Most bacteria contain dozens of TCSs, each of them responsible for sensing and responding to a different range of signals.
The notorious pathogen Staphylococcus aureus contains 16 TCS that are involved in environmental adaptation, disease, and antimicrobial resistance.
Traditionally, the function of each TCS has been determined by analysing the changes in gene expression caused by the absence of individual TCSs.
However, this approach only works when the TCS is actively signalling and cannot resolve TCS regulons that are dormant under the assessed conditions.
In the research, published in mSystems, scientists used an S. aureus strain deprived of the complete TC sensorial system to introduce, one by one, the active form of every TCS avoiding the need for upstream signalling events.
This gain-of-function strategy allowed them to identify the changes in gene expression conferred by each TCS without interference of other members of the family and provides the first description of the complete TCS regulon in a living cell, which they expect will be useful in understanding the pathobiology of this important pathogen.
The project was initially developed by Dr Haag, based in the lab of Professor Jose Penades at the Institute of Infection, Immunity, and Inflammation, with funding obtained from Tenovus Scotland.
Dr Haag said: "In this project, we characterised the complete TCS regulatory network of a bacterial pathogen under identical conditions and without interference from other TCS.
"Our study gives valuable insights into S. aureus TCS-controlled environmental and host adaptation and provides new tools for interrogating the complexity of TCS signal transduction in future studies.
"We are particularly interested in how multiple TCS interact or coordinate to generate cause host adaptation, disease and antimicrobial resistance."
Systematic Reconstruction of the Complete Two-Component Sensorial Network in Staphylococcus aureus
- B. Rapun-Araiz, A. F. Haag, V. De Cesare, C. Gil, P. Dorado-Morales, J. R. Penades, and I. Lasa
- mSystems. 2020 Jul-Aug; 5(4): e00511-20. Published online 2020 Aug 18. doi: 10.1128/mSystems.00511-20
This work was supported by the Spanish Ministry of Economy and Competitiveness, Tenovus Scotland, the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program, the Medical Research Council, and pharmaceutical companies supporting the Division of Signal Transduction Therapy (Boehringer Ingelheim, GlaxoSmithKline, and Merck KGaA).
Image: TCS functional overview. Examples of TCS and their role in S. aureus host adaptation and antimicrobial resistance
First published: 17 September 2020

