Dr Rucha Karnik
- Senior Lecturer (Molecular Biosciences)
telephone:
01413301945
email:
Rucha.Karnik@glasgow.ac.uk
pronouns:
She/her/hers
Bower Building, Room 205, Glasgow, Glasgow City, Scotland, United Kingdom, G12 8SU
Biography
Rucha Karnik comes from an Agri-biotech industry background. She obtained her PhD degree (2011) in Membrane Biology at the University of Leeds with a Overseas Research Students Awards Scheme (ORSAS) studentship award. During her PhD her work focused on indentifying mechanisms for K+ channel traffic impacting human health and disease.
Rucha moved to the University of Glasgow in 2011, taking up a post doctoral position in the Blatt laboratory. Working with plant systems here she focused on elucidating novel mechanisms co-ordinating ion transport and membrane traffic in plants. Rucha was awarded the prestigious Royal Society University Research Fellowship (2016) to establish her own research group at the University of Glasgow and she is now a Senior Royal Society URF.
Research in Karnik lab addresses the overarching question 'How do plants grow?'. Rucha's work is aimed at understanding how plants that we rely on can cope with climate challenges. She investigates the plant molecular machinery involved in multifactorial stress responses impacting plant growth, development and pathogen immunity. Going beyond research in the laboratory, Rucha leads Sci-Seedlets, a cross-disciplinary impact and innovation project to inspire the next generation of plant scientists.
Key words: plant growth, pathogen immunity, membrane transport, stomatal development, stomatal physiology, membrane traffic, climate-change, CO2-sensing, plant blindness
Research interests
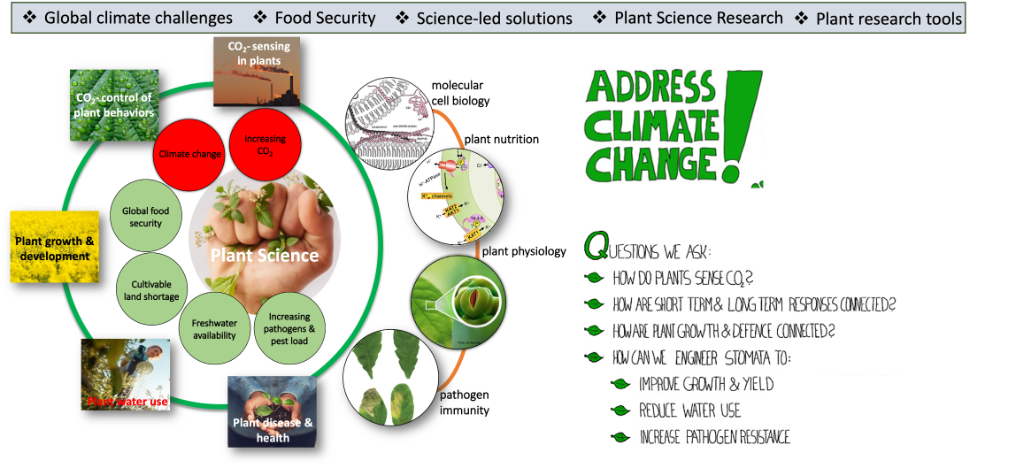
Increasing atmospheric carbon dioxide levels and climate change have a negative impact on crop health and productivity, thus increasing pressure on crop production and limiting the availability of agricultural land and water resources world-wide. Research in Karnik laboratory addresses how do plants grow with a view to understanding how to moderate these pressures. We study molecular mechanisms underpinning immediate responses to multifactorial environmental cues that affect plant growth, immunity and developmental processes. The focus is on stomata, the microscopic pores on plants that facilitate gas exchange for photosynthesis and control plant water loss. We investigate carbon dioxide-sensing mechanisms in plants and their impact on stomatal behaviour. This research crucial for developing fundamental understanding of plant-environment interactions under climate pressure and is an essential first step to ensuring food and water security into the future.
We use multi-disciplinary research platforms including cell biology, protein biochemistry, mass spectrometry, crystallography, live cell imaging, plant physiology and stomatal physiology in our research. Karnik lab research uses model plant Arabidopsis thaliana. Developmental studies are carried out using Begonia species. Research findings are extended to study the horticultural crop cucumber through international collaborations.
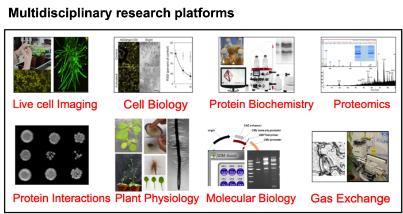
We develop new research tools in molecular biology, protein-protein interaction studies, plant membrane trafficking and plant protein biochemistry to aid research in plant systems.

Current Research Projects
1. Hormone-regulated membrane traffic in plants
Hormonal control of plasma membrane H+-ATPases is mediated in part through trafficking and its co-ordination in response to multifactorial stress. Current interests centre on novel secretory SNARE actions in regulating endocytosis of H+-ATPases.
2. Novel SNARE Partners Facilitating Bacterial Pathogen Defence
Plant microbial pathogens affect plant health inflicting major agricultural and socio-economic losses worldwide. Although plants have evolved defence systems, immunity comes at a cost to plant growth. Microbial pathogens also hijack cellular vesicle traffic to suppress the secretion of defence-related molecules essential for plant immunity. We are interested in the molecular mechanisms regulating of ion and water transport in stomatal guard cells and the integration of pathogen immunity with plant water use.

3. Molecular Mechanisms Underpinning Stomatal Development
Environmental responses of stomata frequently transcend physiological and developmental timescales. We use Begonia species as model to study stomatal development with a view to gain strategic knowledge for development of crops with enhanced stomatal efficiency.
4. Plant Memories and Forgetfulness: Connexions with CO2-sensing and Stomatal responses
Global atmospheric CO2 is predicted to approach levels of 900 μmol mol-1 by the end of the 21st century and is certain to have long-term impacts on stomatal density and patterning on the leaf surface, and consequently on plant biomass. The CO2-sensing mechanisms that integrate stomatal regulation across timescales remain unknown. The laboratory is exploring possible CO2-sensing mechanism that could affect these processes.
Karnik Group and Plant Science at Glasgow
Public Engagement & Innovation

Sci-Seedlets is a cross-disciplinary project to inspire the next generation of plant scientists. We have built a scheme that uses cutting-edge educational formats providing tasters for plant research. Through work in the classroom, Sci-Seedlets supports primary schools, teachers and members of the public with plant science education by exploring topics including 'how do plants grow?', 'how do plants defend against pathogens?' and ‘how to plants respond to their environment?’. Sci-Seedlets resources are now being expanded for use in secondary, undergraduate and postgraduate teaching of plant science.
Find out more!
Sci-Seedlets TRI
Glasgow Science Festival 2022
Royal Society Summer Science Exhibition 2022!
Past Activities
Research groups
Grants
Grants and Awards listed are those received whilst working with the University of Glasgow.
- Structural analysis of a unique K-dependent channel gate
Biotechnology and Biological Sciences Research Council
2025 - 2028
- Cy-BORC - The Vesicle Recycling Complex of Plants
Leverhulme Trust
2025 - 2028
- Sci-Seedlets: Classroom Lesson Kits - building a business model for wider impact
Biotechnology and Biological Sciences Research Council
2025 - 2025
- Plasma Membrane Proton ATPase Cycling in Stress Response
Biotechnology and Biological Sciences Research Council
2024 - 2027
- Dissecting Adaptive Memories in Stomatal Response
Biotechnology and Biological Sciences Research Council
2024 - 2027
- A SNARE-Aquaporin complex in stomatal hydraulics
Biotechnology and Biological Sciences Research Council
2024 - 2026
- Sci-Seedlets - Developing Product Pipeline for Plant Science Education
Biotechnology and Biological Sciences Research Council
2023 - 2025
- IRGA-Live Clamp: An integrated infrared gas-analysis platform to investigate systemic signalling within the plant canopy
Biotechnology and Biological Sciences Research Council
2022 - 2023
- Diversity in STEM
The Royal Society
2022 - 2025
- Plant Memories and Forgetfulness: Connexions with CO2 Perception and Responses
The Royal Society
2022 - 2025
- Sci-Seedlets - BBSRC IAA
Biotechnology and Biological Sciences Research Council
2021 - 2022
- Elucidating Novel Mechanisms For CO2 - Sensing in Plants
Wellcome Trust
2021 - 2022
- Sci-seedlets
The Royal Society
2021 - 2023
- CO2 influence on SNARE regulation
The Royal Society
2021 - 2023
- Sci-Seedlets- Nurturing Plant Science Educational Outcomes in School Children
BBSRC
2021 - 2022
- SYP-SLAC
BBSRC
2021 - 2024
- SNARE endocytosis and secretory vesicle reuse in plant growth-defense trade offs
Biotechnology and Biological Sciences Research Council
2019 - 2022
- How Do Plants Fight Microbes?- The Defence Song
The Royal Society
2019 - 2019
- Proton Transport Modulators - Spatial Regulation and Effects on Plant Physiology
The Royal Society
2017 - 2021
- Hormone-Regulated Membrane Traffic and Plant Morphogenesis
The Royal Society
2016 - 2025
Supervision
PhD Projects
Karnik lab currently hosts PhD students funded by the Royal Society, China Scholarship Council, University of Glasgow and the M L Macintyre Begonia Trust.
Ongoing PhD Projects in the lab:-
1. Engineering Stomtal Efficiency in Plants
*A fully-funded PhD studentship is currently open for applications*
Project Title: Plant CyBORC
Study of novel protein complexes that dictate plant memory to stress.
Fully Funded PhD Studentship 3yrs, on a Leverhulme Trust Funded research grant.
Applications close 4th Feb 2025 (or until filled)
This research project involves cross-disciplinary studies using advanced proteomics, protein-protein interaction analysis and whole plant physiology.
Based in Bower Building, Plant Science Group within a vibrant research environment at the School of Molecular Biosciences, University of Glasgow
Supervisors: Dr Rucha Karnik, Professor Mike Blatt and Dr Sak Waghmare
Informal enquires to Dr Rucha Karnik
Email: Rucha.Karnik@glasgow.ac.uk
Required: UG/ PGT degree in Molecular Biology/ Biochemistry/ Cell Biology or equivalent
Knowledge of Protein Biochemistry is essential.
Interviews of shortlisted candidates: Feb 2025
Self-funded Project
Novel components of the developmental pathways that underpin stomatal clustering.
This PhD programme is to identify . The study will evaluate how stomatal clustering impacts the efficiency of plant growth, water-use and pathogen immunity. The project work will integrate training and research combining cross-disciplinary expertise in genetics, molecular plant pathology, advanced proteomics and stomatal physiology. Knowledge of the fundamental rules of life is anticipated. It will support future crop engineering to mitigate the imminent crisis for agricultural food and freshwater availability and efforts to increased threat for pathogens due to global warming and climate change.
- Ly Truong Phan Thu, *
Engineering stomatal patterning in plants for enhanced water use efficiency
PG & UG Projects
Each year Karnik lab wellcomes undergraduate and post graduate student interns to promote interest in plant scienc. We provide opportunities to engage with our research and contribute to outreach.
Plant Science Internships (Karnik lab)
Summer internship position 2025 - Applications open Feb 2025 (via email to Rucha Karnik). Interviews 1st week of March.
Applied Research Skills Internship 2023 for UofGlasgow students (Supported by Royal Society STEM diversity funding) - 1 position (*now filled)
Summer internships 2023 for UofGlasgow UG students (Supported by Royal Society STEM diversity funding) - 1 position (*now closed*)
Term time internships 2023 for UofGlasgow UG students (Supported by Royal Society STEM diversity funding) - 2 positions (*now closed*)
Summer internship 2023 for UofGlasgow UG Yr1 students - 1 position - *now closed*
Summer School placement 2023 - 1 position - *now closed*
Teaching
-
Level 4: Honours Project Supervisor
-
MSc. Biotechnology assessor & project supervisor
-
MSc. Food Security assessor & project supervisor
-
MSc. Food Security & Management assessor & project supervisor
- MSc Biotechnology BIOL5197 omics course assessor
-
MSc. Food Security, MSc Biotechnology & Level 4 BIOL5312, BIOL5213, BIOL4110 - Plant Research & Biotechnology Platforms
-
Level 3 Biomolecular Sciences Lectures - Membranes & Filaments Module - Vesicle Trafficking Mechanisms, Regulation, Pathophysiology and Plant Growth
- PGR convenor and examiner
Professional activities & recognition
Research fellowships
- 2022 - 2025: Senior Royal Society University Research Fellowship
- 2016 - 2022: Royal Society University Research Fellowship
Grant committees & research advisory boards
- 2020 - 2024: Royal Society, Royal Society Hooke Committee
- 2023 - 2025: Royal Society, Royal Society Grants Committee
- 2024 - 2025: Royal Society, The Future of Scientific Publishing - Advisory Group
- 2024: BBSRC, Pool of Experts
Editorial boards
- 2019 - 2023: Guest Associate Editor for Plant Membrane Traffic and Transport, Frontiers in Plant Science
Professional & learned societies
- 2025: Elected Fellow of the Royal Society of Biology (FRSB), Royal Society of Biology
Selected international presentations
- 2013: European Network for Plant Endomembrane Research (Ghent)
- 2014: Gordon Research Conference: Salt & Water Stress in Plants (Maine NE)
- 2017: European Network for Plant Endomembrane Research (France)
- 2019: European Network for Plant Endomembrane Research (Glasgow)
- 2021: Plant Biology 2021 Worldwide Summit, Cell Biology symposium (virtual)
- 2023: SEB Centenary Conference Edinburgh 2023 (Edinburgh)
- 2024: Royal Society Theo Murphy meeting Carbon dioxide detection in biological systems. (Manchester)



