Dr Brian Smith
- Senior Lecturer (Molecular Biosciences)
telephone:
01413305167
email:
Brian.Smith@glasgow.ac.uk
B4.20 Level B4, School of Molecular Biosciences, Joseph Black Building, Glasgow, G12 8QQ
Research interests
NMR spectroscopy
Nuclear Magnetic Resonance spectroscopy is the only technique that can determine high-resolution structures of macromolecules in solution, and can also probe molecular motion and intermolecular interactions. Brian's group use the technique to study the structures and functions of proteins and nucleic acids involved in interesting processes in a variety of systems. They are also interested in developing NMR methodology. They have a recently updated 600 MHz NMR spectrometer (2013) equipped with a cryoprobe. High-resolution molecular structures are normally determined in vitro, but Brian was part of the team that solved the first structure of a protein inside living cells. We collaborate widely with other biologists and chemists and currently have particular interests in the biophysics and application of natural surfactant proteins, in fatty acid binding proteins from nematode parasites, in the structural basis of epigenetic gene regulation, in the regulation of viral life cycles, in the molecular basis of endotoxin sensing, and in ultraviolet light perception by plants.
Natural surfactant proteins
Surfactants are typically small molecule amphiphiles, but these are typically disruptive to cells. Proteins that act as cell compatible surfactants have arisen independently several times in evolution. Brian's group study ranaspumins found in the foam nests of tropical frogs and the salivary and airway surfactants, equine latherin and human SPLUNC1. The conformational transition that these proteins undergo to convert to their surface active forms sets them apart from other classes of protein surfactants. The group are also exploring biotechnological applications of surfactant proteins.
Collaborators: Prof Malcolm Kennedy, Dr Mathis Riehle, Prof Cait MacPhee (University of Edinburgh), Prof Matt Redinbo (UNC Chapel Hill, USA).
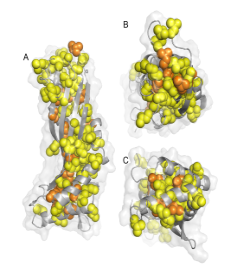
Figure 1 Three views of a cartoon of the structure of the solution form of the surfactant protein latherin showing the locations of the many leucine (yellow) and isoleucine (orange) residues as spheres. (PDB:3ZPM)
Novel fatty acid binding proteins
Brian's group studies the structures and ligand binding properties of novel fatty acid binding proteins from parasites. Parasitic nematodes are responsible for some of the most widespread and pernicious diseases in the developing world as well as causing losses in agriculture. The parasites must scavenge metabolites, including fatty acids and retinoids, from their hosts and maintain a reservoir of fatty acid binding proteins in their pseudocoelemic fluid. Brian's group have determined the structures of nematode polyprotein antigen subunits (NPAs) and fatty acid and retinoid binding proteins (FARs) and more typical FABP proteins and are examining their lipid binding in detail.
Collaborators: Prof Malcolm Kennedy, Prof Betina Córsico (INIBIOLP, Argentina).
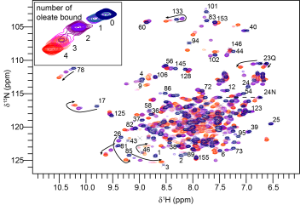
Figure 2 Fatty acid binding by Na-FAR-1 followed by 15N HSQC NMR spectra. The peaks arising from the amide NH of each amino acid residue are seen to jump from one position to another as oleate is titrated into the protein indicating that it can bind multiple ligands, adopting a distinct conformation in each case.
Structural basis of epigenetic gene regulation
Epigenetics describes heritable changes in gene expression that do not involve an alteration of the DNA sequence. For example, as a complex organism develops, its cells differentiate from stem cells into specialised cell types in which groups of genes are down regulated in a heritable fashion. This down regulation requires specialised forms of chromatin (DNA and the proteins that package it) that prevents the transcription machinery from gaining access to the genes. Molecular signals including DNA methylation and modification of histone tails are key to these processes. Brian's group study the structures of the proteins that recognise and interpret these modifications.
Ultraviolet light perception
UVR8, the protein through which plants sense ultraviolet light, is unusual in using tryptophan residues, rather than an additional chromophore. The group are trying to understand the mechanism of UV perception and the conformational changes in the protein that effect downstream signalling.
Collaborators: Prof Gareth Jenkins, Prof John Christie.
Research groups
Publications
2025
Berrocal-Martin, Raul, Sansom, Henry G., Orosz, Katalin, Paterson, Max B., Smith, Brian O. 

Nakasone, Mark, Buetow, Lori, Gabrielsen, Mads 


2024
Dearlove, Emily L., Chatrin, Chatrin, Buetow, Lori, Ahmed, Syed F. 



Pal, Sandeep, Nare, Zandile, Rao, Vincenzo A., Smith, Brian O. 
2023
Adam, Sebastian, Zheng, Dazhong, Klein, Andreas, Volz, Carsten, Mullen, William 


2022
Kowalczyk, Dominika, Nakasone, Mark A., Smith, Brian O. 

Gracie, Jennifer, Zamberlan, Francesco, Andrews, Iain B., Smith, Brian O. 

Ryan, S. M. et al. (2022) Novel antiinflammatory biologics shaped by parasite–host coevolution. Proceedings of the National Academy of Sciences of the United States of America, 119(36), e220279511. (doi: 10.1073/pnas.2202795119) (PMID:36037362)
Kjeldsen, A. et al. (2022) The fluorescent protein iLOV as a reporter for screening of high‐yield production of antimicrobial peptides in Pichia pastoris. Microbial Biotechnology, 15(7), pp. 2126-2139. (doi: 10.1111/1751-7915.14034) (PMID:35312165) (PMCID:PMC9249318)
Torres Cabán, Cristina C., Yang, Minghan, Lai, Cuixin, Yang, Lina, Subach, Fedor V., Smith, Brian O. 
Tibbo, A. J. et al. (2022) Phosphodiesterase type 4 anchoring regulates cAMP signaling to Popeye domain-containing proteins. Journal of Molecular and Cellular Cardiology, 165, pp. 86-102. (doi: 10.1016/j.yjmcc.2022.01.001) (PMID:34999055) (PMCID:PMC8986152)
Nakasone, Mark A., Majorek, Karolina A., Gabrielsen, Mads 


Neckebroeck, Albane, Kelly, Sharon M. 


2021
Prabhakar, A. T. et al. (2021) CK2 phosphorylation of human papillomavirus 16 E2 on serine 23 promotes interaction with TopBP1 and is critical for E2 interaction with mitotic chromatin and the viral life cycle. mBio, 12, e01163-21. (doi: 10.1128/mbio.01163-21) (PMID:34544280) (PMCID:PMC8546539)
Buist, H. K. et al. (2021) Identification and characterization of an affimer affinity reagent for the detection of the cAMP sensor, EPAC1. Cells, 10(9), 2307. (doi: 10.3390/cells10092307)
Morgan, D. C. et al. (2021) Stapled ACE2 peptidomimetics designed to target the SARS-CoV-2 spike protein do not prevent virus internalisation. Peptide Science, 113(4), e24217. (doi: 10.1002/pep2.24217) (PMID:33615115) (PMCID:PMC7883042)
Knuhtsen, Astrid, Whiting, Rachel, McWhinnie, Fergus S., Whitmore, Charlotte, Smith, Brian O. 

2020
Chatrin, Chatrin, Gabrielsen, Mads 


Crecente Garcia, Selma, Neckebroeck, Albane, Clark, J. Stephen 


2019
Ibáñez-Shimabukuro, M. et al. (2019) Structure and ligand binding of As-p18, an extracellular fatty acid binding protein from the eggs of a parasitic nematode. Bioscience Reports, 39(7), BSR2019129. (doi: 10.1042/BSR20191292) (PMID:31273060) (PMCID:PMC6646235)
Knuhtsen, Astrid, Whitmore, Charlotte, McWhinnie, Fergus S., McDougall, Laura, Whiting, Rachel, Smith, Brian O. 

2017
Cooper, Alan, Vance, Steven J., Smith, Brian O. 

Gabrielsen, Mads 


Magennis, Steve 


Spies, Maria and Smith, Brian O. 
Brandani, Giovanni B., Vance, Steven J., Schor, Marieke, Cooper, Alan, Kennedy, Malcolm W. 

Parnell, Euan, McElroy, Stuart P., Wiejak, Jolanta, Baillie, Gemma L., Porter, Alison, Adams, David R., Rehmann, Holger, Smith, Brian O. 
2016
Grinter, R. et al. (2016) Structure of the bacterial plant-ferredoxin receptor FusA. Nature Communications, 7, 13308. (doi: 10.1038/ncomms13308) (PMID:27796364) (PMCID:PMC5095587)
Heilmann, Monika, Velanis, Christos N., Cloix, Catherine, Smith, Brian O. 


Morris, Ryan J., Brandani, Giovanni B., Desai, Vibhuti, Smith, Brian O. 
Vance, Steven J., Desai, Vibhuti, Smith, Brian O. 

Ashraf, Khuram U., Josts, Inokentijs, Mosbahi, Khedidja, Kelly, Sharon M. 



2015
Rey, B. M.F. et al. (2015) Diversity in the structures and ligand binding sites of nematode fatty acid and retinol binding proteins revealed by Na-FAR-1 from Necator americanus. Biochemical Journal, 471(3), pp. 403-414. (doi: 10.1042/BJ20150068) (PMID:26318523) (PMCID:PMC4613501)
Thomson, Ross and Smith, Brian O. 
Parnell, Euan, Smith, Brian O. 
Buetow, Lori, Gabrielsen, Mads 


Shigemitsu, Yoshiki, Ikeya, Teppei, Yamamoto, Akihiro, Tsuchie, Yuusuke, Mishima, Masaki, Smith, Brian O. 
Franchini, Gisela R., Pórfido, Jorge L., Ibáñez Shimabukuro, Marina, Rey Burusco, María F., Bélgamo, Julián A., Smith, Brian O. 

Mathes, T. et al. (2015) Proton-coupled electron transfer constitutes the photoactivation mechanism of the plant photoreceptor UVR8. Journal of the American Chemical Society, 137(25), pp. 8113-8120. (doi: 10.1021/jacs.5b01177) (PMID:25955727)
2014
Beckham, K. S.H. et al. (2014) The metabolic enzyme AdhE controls the virulence of Escherichia coli O157:H7. Molecular Microbiology, 93(1), pp. 199-211. (doi: 10.1111/mmi.12651) (PMID:24846743) (PMCID:PMC4249723)
Cameron, Ryan T., Quinn, Steven D. 


Ibáñez-Shimabukuro, Marina, Rey-Burusco, M.Florencia, Cooper, Alan, Kennedy, Malcolm W. 

Vance, Steven J., McDonald, Rhona E., Cooper, Alan, Kennedy, Malcolm W. 

Yvon, Carine, Surman, Andrew J. 



Rey-Burusco, M.Florencia, Ibañez-Shimabukuro, Marina, Cooper, Alan, Kennedy, Malcolm W. 

McCaughey, L. C. et al. (2014) Lectin-like bacteriocins from Pseudomonas spp. utilise D-rhamnose containing lipopolysaccharide as a cellular receptor. PLoS Pathogens, 10(2), e1003898. (doi: 10.1371/journal.ppat.1003898) (PMID:24516380) (PMCID:PMC3916391)
Caldwell, S.T. et al. (2014) Protein-mediated dethreading of a biotin-functionalised pseudorotaxane. Organic and Biomolecular Chemistry, 12(3), pp. 511-516. (doi: 10.1039/C3OB41612G) (PMID:24280954)
Josts, Inokentijs, Grinter, Rhys, Kelly, Sharon M. 




2013
Laird, J. et al. (2013) Identification of the domains of cauliflower mosaic virus protein P6 responsible for suppression of RNA silencing and salicylic acid signalling. Journal of General Virology, 94(12), pp. 2777-2789. (doi: 10.1099/vir.0.057729-0) (PMID:24088344) (PMCID:PMC3836500)
Hamatsu, J. et al. (2013) High-resolution heteronuclear multidimensional NMR of proteins in living insect cells using a baculovirus protein expression system. Journal of the American Chemical Society, 135(5), pp. 1688-1691. (doi: 10.1021/ja310928u)
Kennedy, M.W., Córsico, B., Cooper, A. and Smith, B.O. (2013) The unusual lipid-binding proteins of nematodes: NPAs, nemFABPs and FARs. In: Kennedy, M. and Harnett, m. (eds.) Parasitic Nematodes: Molecular Biology, Biochemistry and Immunology [2nd. ed.]. CABI; Wallingford; UK, pp. 397-412. ISBN 9781845937591 (doi: 10.1079/9781845937591.0397)
Vance, S.J, McDonald, R.E, Cooper, A., Smith, B.O. 

2012
Cloix, C., Kaiserli, E. 




Gabrielsen, M. 



Parnell, Euan, Smith, Brian O. 
Christie, J.M. et al. (2012) Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science, 335(6075), pp. 1492-1496. (doi: 10.1126/science.1218091)
Gabrielsen, M. 



Gabrielsen, M. 



2011
Wang, D. et al. (2011) Identification of bacterial target proteins for the salicylidene acylhydrazide class of virulence blocking compounds. Journal of Biological Chemistry, 286(34), pp. 29922-29931. (doi: 10.1074/jbc.M111.233858) (PMID:21724850) (PMCID:PMC3191033)
Meenan, N.A.G., Ball, G., Bromek, K., Uhrín, D., Cooper, A., Kennedy, M.W. 

2010
Ikeya, T. et al. (2010) NMR protein structure determination in living E. coli cells using nonlinear sampling. Nature Protocols, 5(6), pp. 1051-1060. (doi: 10.1038/nprot.2010.69)
2009
MacKenzie, C.D., Smith, B.O., Meister, A., Blume, A., Zhao, X., Lu, J.R., Kennedy, M.W. and Cooper, A. (2009) Ranaspumin-2: structure and function of a surfactant protein from the foam nests of a tropical frog. Biophysical Journal, 96(12), pp. 4984-4992. (doi: 10.1016/j.bpj.2009.03.044)
Sakakibara, D. et al. (2009) Protein structure determination in living cells by in-cell NMR spectroscopy. Nature, 458(7234), pp. 102-105. (doi: 10.1038/nature07814)
Borland, G., Smith, B.O. and Yarwood, S.J. (2009) EPAC proteins transduce diverse cellular actions of cAMP. British Journal of Pharmacology, 158(1), pp. 70-86. (doi: 10.1111/j.1476-5381.2008.00087.x)
2008
dos Reis, Flavia C.G., Smith, Brian O., Santos, Camila C., Costa, Tatiana, F.R., Scharfstein, Julio, Coombs, Graham H., Mottram, Jeremy C. and Lima, Ana Paula C.A. (2008) The role of conserved residues of chagasin in the inhibition of cysteine peptidases. FEBS Letters, 582(4), pp. 485-490. (doi: 10.1016/j.febslet.2008.01.008)
Caldwell, S. T. 


Caldwell, S. T. 


2007
Alexander, A. et al. (2007) Probing the solvent-induced tautomerism of a redox-active ureidopyrimidinone. Chemical Communications, pp. 2246-2248. (doi: 10.1039/b703070c)
2006
Smith, B.O., Picken, N.C., Westrop, G.D., Bromek, K., Mottram, J.C. 
Ball, G, Meenan, N, Bromek, K, Smith, BO, Bella, J and Uhrin, D (2006) Measurement of one-bond C-13(alpha)-H-1(alpha) residual dipolar coupling constants in proteins by selective manipulation of (CH alpha)-H-alpha spins. Journal of Magnetic Resonance, 180, pp. 127-136. (doi: 10.1016/j.jmr.2006.01.017)
Smith, B.O., Westrop, G.D., Mottram, J.C. and Coombs, G.H. (2006) Chemical shift assignments of Leishmania mexicana ICP, a novel cysteine peptidase inhibitor. Journal of Biomolecular NMR, 36, p. 7. (doi: 10.1007/s10858-005-4739-8)
2005
Meenan, N.A.G., Cooper, A., Kennedy, M.W. 

2004
Weber, M.A. et al. (2004) Blood pressure dependent and independent effects of antihypertensive treatment on clinical events in the VALUE Trial. Lancet, 363(9426), pp. 2049-2051. (doi: 10.1016/S0140-6736(04)16456-8)
Smith, B (2004) Structural analysis of the complement control protein (CCP) modules of GABAB receptor 1a: only one of the two ccp modules is compactly folded. Journal of Biological Chemistry, 279(46), pp. 48292-48306. (doi: 10.1074/jbc.M406540200)
2002
Bramham, J, Hodgkinson, JL, Smith, BO, Uhrin, D, Barlow, PN and Winder, SJ (2002) Solution structure of the calponin CH domain and fitting to the 3D-helical reconstruction of F-actin : calponin. Structure, 10, pp. 249-258.
Smith, B (2002) Structure of the C3b binding site of CR1 (CD35), the immune adherence receptor. Cell, 108(6), pp. 769-780. (doi: 10.1016/S0092-8674(02)00672-4)
2001
Smith, B (2001) DNA recognition by the methyl-CpG binding domain of MeCP2. Journal of Biological Chemistry, 276(5), pp. 3353-3360. (doi: 10.1074/jbc.M007224200)
Smith, B (2001) Solution structure and dynamics of the central CCP module pair of a poxvirus complement control protein. Journal of Molecular Biology, 307(1), pp. 323-339. (doi: 10.1006/jmbi.2000.4477)
Articles
Berrocal-Martin, Raul, Sansom, Henry G., Orosz, Katalin, Paterson, Max B., Smith, Brian O. 

Nakasone, Mark, Buetow, Lori, Gabrielsen, Mads 


Dearlove, Emily L., Chatrin, Chatrin, Buetow, Lori, Ahmed, Syed F. 



Pal, Sandeep, Nare, Zandile, Rao, Vincenzo A., Smith, Brian O. 
Adam, Sebastian, Zheng, Dazhong, Klein, Andreas, Volz, Carsten, Mullen, William 


Kowalczyk, Dominika, Nakasone, Mark A., Smith, Brian O. 

Gracie, Jennifer, Zamberlan, Francesco, Andrews, Iain B., Smith, Brian O. 

Ryan, S. M. et al. (2022) Novel antiinflammatory biologics shaped by parasite–host coevolution. Proceedings of the National Academy of Sciences of the United States of America, 119(36), e220279511. (doi: 10.1073/pnas.2202795119) (PMID:36037362)
Kjeldsen, A. et al. (2022) The fluorescent protein iLOV as a reporter for screening of high‐yield production of antimicrobial peptides in Pichia pastoris. Microbial Biotechnology, 15(7), pp. 2126-2139. (doi: 10.1111/1751-7915.14034) (PMID:35312165) (PMCID:PMC9249318)
Torres Cabán, Cristina C., Yang, Minghan, Lai, Cuixin, Yang, Lina, Subach, Fedor V., Smith, Brian O. 
Tibbo, A. J. et al. (2022) Phosphodiesterase type 4 anchoring regulates cAMP signaling to Popeye domain-containing proteins. Journal of Molecular and Cellular Cardiology, 165, pp. 86-102. (doi: 10.1016/j.yjmcc.2022.01.001) (PMID:34999055) (PMCID:PMC8986152)
Nakasone, Mark A., Majorek, Karolina A., Gabrielsen, Mads 


Neckebroeck, Albane, Kelly, Sharon M. 


Prabhakar, A. T. et al. (2021) CK2 phosphorylation of human papillomavirus 16 E2 on serine 23 promotes interaction with TopBP1 and is critical for E2 interaction with mitotic chromatin and the viral life cycle. mBio, 12, e01163-21. (doi: 10.1128/mbio.01163-21) (PMID:34544280) (PMCID:PMC8546539)
Buist, H. K. et al. (2021) Identification and characterization of an affimer affinity reagent for the detection of the cAMP sensor, EPAC1. Cells, 10(9), 2307. (doi: 10.3390/cells10092307)
Morgan, D. C. et al. (2021) Stapled ACE2 peptidomimetics designed to target the SARS-CoV-2 spike protein do not prevent virus internalisation. Peptide Science, 113(4), e24217. (doi: 10.1002/pep2.24217) (PMID:33615115) (PMCID:PMC7883042)
Knuhtsen, Astrid, Whiting, Rachel, McWhinnie, Fergus S., Whitmore, Charlotte, Smith, Brian O. 

Chatrin, Chatrin, Gabrielsen, Mads 


Crecente Garcia, Selma, Neckebroeck, Albane, Clark, J. Stephen 


Ibáñez-Shimabukuro, M. et al. (2019) Structure and ligand binding of As-p18, an extracellular fatty acid binding protein from the eggs of a parasitic nematode. Bioscience Reports, 39(7), BSR2019129. (doi: 10.1042/BSR20191292) (PMID:31273060) (PMCID:PMC6646235)
Knuhtsen, Astrid, Whitmore, Charlotte, McWhinnie, Fergus S., McDougall, Laura, Whiting, Rachel, Smith, Brian O. 

Cooper, Alan, Vance, Steven J., Smith, Brian O. 

Gabrielsen, Mads 


Magennis, Steve 


Spies, Maria and Smith, Brian O. 
Brandani, Giovanni B., Vance, Steven J., Schor, Marieke, Cooper, Alan, Kennedy, Malcolm W. 

Parnell, Euan, McElroy, Stuart P., Wiejak, Jolanta, Baillie, Gemma L., Porter, Alison, Adams, David R., Rehmann, Holger, Smith, Brian O. 
Grinter, R. et al. (2016) Structure of the bacterial plant-ferredoxin receptor FusA. Nature Communications, 7, 13308. (doi: 10.1038/ncomms13308) (PMID:27796364) (PMCID:PMC5095587)
Heilmann, Monika, Velanis, Christos N., Cloix, Catherine, Smith, Brian O. 


Morris, Ryan J., Brandani, Giovanni B., Desai, Vibhuti, Smith, Brian O. 
Vance, Steven J., Desai, Vibhuti, Smith, Brian O. 

Ashraf, Khuram U., Josts, Inokentijs, Mosbahi, Khedidja, Kelly, Sharon M. 



Rey, B. M.F. et al. (2015) Diversity in the structures and ligand binding sites of nematode fatty acid and retinol binding proteins revealed by Na-FAR-1 from Necator americanus. Biochemical Journal, 471(3), pp. 403-414. (doi: 10.1042/BJ20150068) (PMID:26318523) (PMCID:PMC4613501)
Thomson, Ross and Smith, Brian O. 
Parnell, Euan, Smith, Brian O. 
Buetow, Lori, Gabrielsen, Mads 


Shigemitsu, Yoshiki, Ikeya, Teppei, Yamamoto, Akihiro, Tsuchie, Yuusuke, Mishima, Masaki, Smith, Brian O. 
Franchini, Gisela R., Pórfido, Jorge L., Ibáñez Shimabukuro, Marina, Rey Burusco, María F., Bélgamo, Julián A., Smith, Brian O. 

Mathes, T. et al. (2015) Proton-coupled electron transfer constitutes the photoactivation mechanism of the plant photoreceptor UVR8. Journal of the American Chemical Society, 137(25), pp. 8113-8120. (doi: 10.1021/jacs.5b01177) (PMID:25955727)
Beckham, K. S.H. et al. (2014) The metabolic enzyme AdhE controls the virulence of Escherichia coli O157:H7. Molecular Microbiology, 93(1), pp. 199-211. (doi: 10.1111/mmi.12651) (PMID:24846743) (PMCID:PMC4249723)
Cameron, Ryan T., Quinn, Steven D. 


Ibáñez-Shimabukuro, Marina, Rey-Burusco, M.Florencia, Cooper, Alan, Kennedy, Malcolm W. 

Vance, Steven J., McDonald, Rhona E., Cooper, Alan, Kennedy, Malcolm W. 

Yvon, Carine, Surman, Andrew J. 



Rey-Burusco, M.Florencia, Ibañez-Shimabukuro, Marina, Cooper, Alan, Kennedy, Malcolm W. 

McCaughey, L. C. et al. (2014) Lectin-like bacteriocins from Pseudomonas spp. utilise D-rhamnose containing lipopolysaccharide as a cellular receptor. PLoS Pathogens, 10(2), e1003898. (doi: 10.1371/journal.ppat.1003898) (PMID:24516380) (PMCID:PMC3916391)
Caldwell, S.T. et al. (2014) Protein-mediated dethreading of a biotin-functionalised pseudorotaxane. Organic and Biomolecular Chemistry, 12(3), pp. 511-516. (doi: 10.1039/C3OB41612G) (PMID:24280954)
Josts, Inokentijs, Grinter, Rhys, Kelly, Sharon M. 




Laird, J. et al. (2013) Identification of the domains of cauliflower mosaic virus protein P6 responsible for suppression of RNA silencing and salicylic acid signalling. Journal of General Virology, 94(12), pp. 2777-2789. (doi: 10.1099/vir.0.057729-0) (PMID:24088344) (PMCID:PMC3836500)
Hamatsu, J. et al. (2013) High-resolution heteronuclear multidimensional NMR of proteins in living insect cells using a baculovirus protein expression system. Journal of the American Chemical Society, 135(5), pp. 1688-1691. (doi: 10.1021/ja310928u)
Vance, S.J, McDonald, R.E, Cooper, A., Smith, B.O. 

Cloix, C., Kaiserli, E. 




Gabrielsen, M. 



Parnell, Euan, Smith, Brian O. 
Christie, J.M. et al. (2012) Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science, 335(6075), pp. 1492-1496. (doi: 10.1126/science.1218091)
Gabrielsen, M. 



Gabrielsen, M. 



Wang, D. et al. (2011) Identification of bacterial target proteins for the salicylidene acylhydrazide class of virulence blocking compounds. Journal of Biological Chemistry, 286(34), pp. 29922-29931. (doi: 10.1074/jbc.M111.233858) (PMID:21724850) (PMCID:PMC3191033)
Meenan, N.A.G., Ball, G., Bromek, K., Uhrín, D., Cooper, A., Kennedy, M.W. 

Ikeya, T. et al. (2010) NMR protein structure determination in living E. coli cells using nonlinear sampling. Nature Protocols, 5(6), pp. 1051-1060. (doi: 10.1038/nprot.2010.69)
MacKenzie, C.D., Smith, B.O., Meister, A., Blume, A., Zhao, X., Lu, J.R., Kennedy, M.W. and Cooper, A. (2009) Ranaspumin-2: structure and function of a surfactant protein from the foam nests of a tropical frog. Biophysical Journal, 96(12), pp. 4984-4992. (doi: 10.1016/j.bpj.2009.03.044)
Sakakibara, D. et al. (2009) Protein structure determination in living cells by in-cell NMR spectroscopy. Nature, 458(7234), pp. 102-105. (doi: 10.1038/nature07814)
Borland, G., Smith, B.O. and Yarwood, S.J. (2009) EPAC proteins transduce diverse cellular actions of cAMP. British Journal of Pharmacology, 158(1), pp. 70-86. (doi: 10.1111/j.1476-5381.2008.00087.x)
dos Reis, Flavia C.G., Smith, Brian O., Santos, Camila C., Costa, Tatiana, F.R., Scharfstein, Julio, Coombs, Graham H., Mottram, Jeremy C. and Lima, Ana Paula C.A. (2008) The role of conserved residues of chagasin in the inhibition of cysteine peptidases. FEBS Letters, 582(4), pp. 485-490. (doi: 10.1016/j.febslet.2008.01.008)
Caldwell, S. T. 


Caldwell, S. T. 


Alexander, A. et al. (2007) Probing the solvent-induced tautomerism of a redox-active ureidopyrimidinone. Chemical Communications, pp. 2246-2248. (doi: 10.1039/b703070c)
Smith, B.O., Picken, N.C., Westrop, G.D., Bromek, K., Mottram, J.C. 
Ball, G, Meenan, N, Bromek, K, Smith, BO, Bella, J and Uhrin, D (2006) Measurement of one-bond C-13(alpha)-H-1(alpha) residual dipolar coupling constants in proteins by selective manipulation of (CH alpha)-H-alpha spins. Journal of Magnetic Resonance, 180, pp. 127-136. (doi: 10.1016/j.jmr.2006.01.017)
Smith, B.O., Westrop, G.D., Mottram, J.C. and Coombs, G.H. (2006) Chemical shift assignments of Leishmania mexicana ICP, a novel cysteine peptidase inhibitor. Journal of Biomolecular NMR, 36, p. 7. (doi: 10.1007/s10858-005-4739-8)
Meenan, N.A.G., Cooper, A., Kennedy, M.W. 

Weber, M.A. et al. (2004) Blood pressure dependent and independent effects of antihypertensive treatment on clinical events in the VALUE Trial. Lancet, 363(9426), pp. 2049-2051. (doi: 10.1016/S0140-6736(04)16456-8)
Smith, B (2004) Structural analysis of the complement control protein (CCP) modules of GABAB receptor 1a: only one of the two ccp modules is compactly folded. Journal of Biological Chemistry, 279(46), pp. 48292-48306. (doi: 10.1074/jbc.M406540200)
Bramham, J, Hodgkinson, JL, Smith, BO, Uhrin, D, Barlow, PN and Winder, SJ (2002) Solution structure of the calponin CH domain and fitting to the 3D-helical reconstruction of F-actin : calponin. Structure, 10, pp. 249-258.
Smith, B (2002) Structure of the C3b binding site of CR1 (CD35), the immune adherence receptor. Cell, 108(6), pp. 769-780. (doi: 10.1016/S0092-8674(02)00672-4)
Smith, B (2001) DNA recognition by the methyl-CpG binding domain of MeCP2. Journal of Biological Chemistry, 276(5), pp. 3353-3360. (doi: 10.1074/jbc.M007224200)
Smith, B (2001) Solution structure and dynamics of the central CCP module pair of a poxvirus complement control protein. Journal of Molecular Biology, 307(1), pp. 323-339. (doi: 10.1006/jmbi.2000.4477)
Book Sections
Kennedy, M.W., Córsico, B., Cooper, A. and Smith, B.O. (2013) The unusual lipid-binding proteins of nematodes: NPAs, nemFABPs and FARs. In: Kennedy, M. and Harnett, m. (eds.) Parasitic Nematodes: Molecular Biology, Biochemistry and Immunology [2nd. ed.]. CABI; Wallingford; UK, pp. 397-412. ISBN 9781845937591 (doi: 10.1079/9781845937591.0397)
Grants
Grants and Awards listed are those received whilst working with the University of Glasgow.
- Disarming Type 3 Secretion - elucidating the mechanism of action for aurodox
Biotechnology and Biological Sciences Research Council
2024 - 2027
- Facilitate the use of NMR spectroscopy by scientists based in Scotland
University of Edinburgh
2022 - 2023
- The Collaborative Computational Project for NMR (CCPN): data analysis, integration and partnerships for NMR in the biosciences
Medical Research Council
2021 - 2026
- Co-ordinated Photoreceptor Engineering for Improved Biomass Production
Biotechnology and Biological Sciences Research Council
2020 - 2023
- Role of reversible modification of methionine residues in the regulation of protein function
Biotechnology and Biological Sciences Research Council
2020 - 2023
- Scottish High-Field NMR Facility
Engineering and Physical Sciences Research Council (EPSRC)
2018 - 2021
- A novel mechanism of protein uptake in Gram-negative bacteria
Biotechnology and Biological Sciences Research Council
2014 - 2018
- Assessing the predictive value of quantitative high-throughput NMR metabolomic analysis for CVD events in a major study of diabetes: ADVANCE
Chest Heart and Stroke Scotland
2013 - 2016
- How does the plant UV-B photoreceptor UVR8 initiate signalling?
Biotechnology and Biological Sciences Research Council
2013 - 2016
- Final structure refinement and functional analysis of Na-FAR-1
Boehringer Ingelheim
2012 - 2012
- Structure determination and functional analysis through Nuclear Magnetic Resonance
Boehringer Ingelheim
2012 - 2012
- The application of a high-throughput NMR metabolomics system to the study of insulin resistance (ISSF Catalyst)
Wellcome Trust
2011 - 2014
- Interaction between the essential replication factor TopBP1 and human papillomavirus 16 E1: a novel antiviral target
Cancer Research UK
2011 - 2015
- Identification of small molecule activators of EPAC1 to serve as novel anti-inflamatory agents in vascular endothelial cells
Scottish Universities Life Sciences Alliance
2011 - 2012
- Quest for the Holy Grail of plant photobiology: a UV-B photoreceptor
Leverhulme Trust
2010 - 2014
- Structural and biophysical analysis of novel lipid binding proteins from parasitic helminths
Wellcome Trust
2008 - 2012
- The solution structures of newly discovered natural inhibitors of cystine peptidases from eukaryotic and bacterial pathogens
Wellcome Trust
2004 - 2008
- Toward Understanding the structural basis of molecular recognition by methylated DNA binding proteins
The Royal Society
2004 - 2005
- Establishment of high field NMR facility
Wellcome Trust
2003 - 2006
Research datasets
2017
Gabrielsen, M., Buetow, L., Nakasone, M., Ahmed, S., Sibbet, G., Smith, B. , Zhang, W., Sidhu, S. and Huang, D. (2017) A general strategy for discovery of inhibitors and activators of RING and U-box E3 ligases with ubiquitin variants. [Data Collection]
2015
Buetow, L., Gabrielsen, M., Anthony, N., Dou, H., Patel, A., Aitkenhead, H., Sibbet, G., Smith, B. and Huang, D. (2015) Activation of a primed RING E3-E2-ubiquitn complex by non-covalent ubiquitin. [Data Collection]


