Dissecting immune cell accumulation in the inflamed joint
Published: 10 March 2022
Institute scientists have used a combination of novel cell tracking approaches and murine models of inflammatory arthritis to identify genes, pathways and anatomically-specific tissue signatures regulating cell migration in a variety of inflamed sites to provide an invaluable resource for the identification of novel therapeutic targets for arthritis and other inflammatory disorders.
Institute scientists have used a combination of novel cell tracking approaches and murine models of inflammatory arthritis to identify genes, pathways and anatomically-specific tissue signatures regulating cell migration in a variety of inflamed sites.
This unique skin and joint specific dataset will be an invaluable resource for the identification of novel therapeutic targets for arthritis and other inflammatory disorders.
Cells of the immune system accumulate in the tissues of the body in autoimmune and inflammatory conditions such as rheumatoid arthritis and once there can cause damage, which may lead to loss of structural integrity and function.
This accumulation may be a consequence of increased numbers of cells entering and/or decreased exit. However, mechanisms governing entry and exit of immune cells into, and out of, inflamed joints, remain poorly understood.
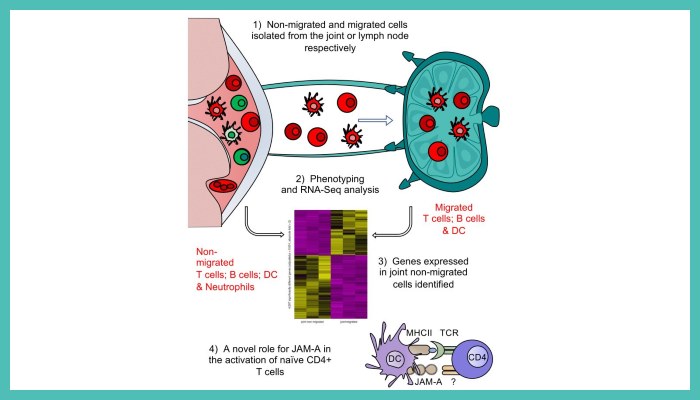
In this work, published in JCI Insight, researchers sought to identify the key molecular pathways regulating such migration.
Using murine models of inflammation in conjunction with mice expressing a photoconvertible fluorescent protein, they characterised the migration of cells from joints to draining lymph nodes (LN) and performed RNA-seq analysis on isolated cells, identifying genes associated with migration and retention.
Following further refining of the gene list to those specific for joint inflammation, RNA-seq data revealed pathways and genes previously highlighted as characteristic of RA in patient studies, validating the methodology.
Focusing on gene regulatory pathways associated with cell migration, adhesion and movement, they identified genes involved in the retention of immune cells in the inflamed joint, namely JAM-A, and identified a role for such molecules in T cell differentiation in vivo.
Thus, Institute scientists have used imaging and molecular approaches to ‘tag' cells in different types of inflamed tissues, follow those that stay or leave, and determine some of the molecules that control these processes.
Professor Paul Garside said: "Using a combination of imaging and molecular techniques, we have identified anatomically specific genetic signatures controlling accumulation of immune cells in different types of target tissues.
"By increasing our understanding of the molecular processes that control cell migration in the context of autoimmune and inflammatory disease, we hope to better target existing therapies and develop novel therapeutic approaches in the future."
Dissecting the molecular control of immune cell accumulation in the inflamed joint
- Catriona T. Prendergast, Robert A. Benson, Hannah E. Scales, Caio S. Bonilha, John J. Cole, Iain McInnes, James M. Brewer, and Paul Garside
- Published: 22 February 2022. Online ahead of print. PMID: 35192549 Free article.
- JCI Insight. doi: 10.1172/jci.insight.151281.
Image: Graphical Abstract, by Dr Hannah Scales
First published: 10 March 2022

