Machine learning approaches classify malaria outcomes
Published: 10 December 2020
A study involving the Institute's Dr Thomas Otto has provided proof of concept for methods that classify uncomplicated malaria and severe malaria from non-malarial infections, showing that the machine learning approach is a feasible tool for clinical decision support.
A research article produced by an international collaboration involving the Institute's Dr Thomas Otto and colleagues in Ghana, Sheffield and London was recently published by BMC Medicine.
The study aimed to evaluate machine learning approaches that can accurately classify non-malarial infections (nMI), uncomplicated malaria, and severe malaria (SM) using haematological parameters as a new application for precision medicine.
Malaria is still a major global health burden, with more than 3.2 billion people in 91 countries remaining at risk of the disease.
Accurately distinguishing malaria from other diseases, especially uncomplicated malaria from non-malarial infections remains a challenge.
Six different machine learning approaches were tested, to select the best approach. An artificial neural network (ANN) with three hidden layers was used for multi-classification of UM, SM, and uMI.
Binary classifiers were developed to further identify the parameters that can distinguish UM or SM from nMI. Local interpretable model-agnostic explanations (LIME) were used to explain the binary classifiers.
The results showed that the multi-classification model had greater than 85 per cent training and testing accuracy to distinguish clinical malaria from non-malarial infections.
To distinguish UM from nMI, their approach identified platelet counts, red blood cell (RBC) counts, lymphocyte counts, and percentages as the top classifiers of UM with 0.801 test accuracy (AUC = 0.866 and F1 score = 0.747).
To distinguish SM from nMI, the classifier had a test accuracy of 0.96 (AUC = 0.983 and F1 score = 0.944) with mean platelet volume and mean cell volume being the unique classifiers of SM.
Random forest was used to confirm the classifications, and it showed that platelet and RBC counts were the major classifiers of UM, regardless of possible confounders such as patient age and sampling location.
Overall, the study provides proof of concept methods that classify uncomplicated malaria and severe malaria from non-malarial infections, showing that the machine learning approach is a feasible tool for clinical decision support.
In the future, machine learning approaches could be incorporated into clinical decision-support algorithms for the diagnosis of acute febrile illness and monitoring response to acute SM treatment particularly in endemic settings.
Machine learning approaches classify clinical malaria outcomes based on haematological parameters
- Collins M. Morang’a, Lucas Amenga–Etego, Saikou Y. Bah, Vincent Appiah, Dominic S. Y. Amuzu, Nicholas Amoako, James Abugri, Abraham R. Oduro, Aubrey J. Cunnington, Gordon A. Awandare and Thomas D. Otto
- BMC Medicine (2020) 18:375 https://doi.org/10.1186/s12916-020-01823-3
Funding: The study was supported by a DELTAS Africa grant (DEL-15-007: Awandare). The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS)’s Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust and the UK government. TO is supported by the Wellcome Trust grant.
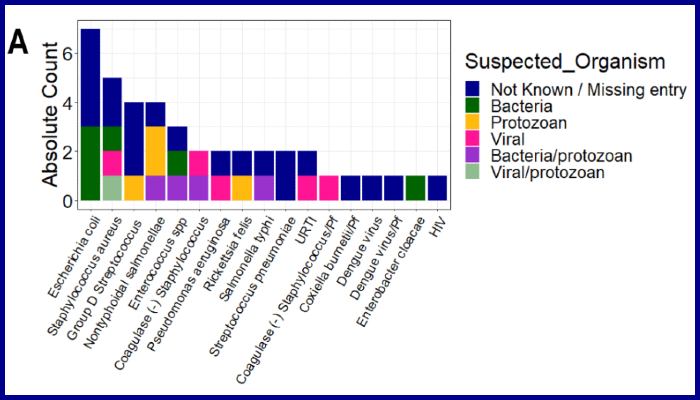
Image legend (Figure 2a): Clinical manifestations using laboratory diagnosis compared to various suspected infections by clinicians. Blood, urine, and stool samples were collected from majority of the individuals who were categorized as nMI. Cultures of either blood, urine, or stool were performed, depending on the clinician’s request and the suspected illness. The suspected organisms were categorized as either bacteria, viral, fungi, and protozoan or a combination of bacteria/protozoan, fungi/protozoan, viral/protozoan, and viral/bacteria. Laboratory results confirmed only 4% of the cases with the majority being undetermined/not available/not known (96%, 937/978). The major organisms determined to be present include dengue virus, Staphylococcus aureus, Salmonella typhi, Streptococcus pneumonia, and Coxiella burnetii. a shows the absolute counts of each diagnosed organism coloured by the suspected organisms.
First published: 10 December 2020
Related Links
- Dr Thomas Otto profile
- West African Centre for Cell Biology of Infectious Pathogens (WACCBIP)
- University of Ghana
- Florey Institute
- University of Sheffield
- C. K. Tedam University of Technology and Applied Sciences
- Navrongo Health Research Centre (NHRC)
- Department of Infectious Disease, Imperial College London
- BMC Medicine
- Read the paper in BMC Medicine


