Roe Research group
This is the webpage of Andrew J. Roe's research group.
The major focus of our work is the model organism, Escherichia coli, which is very diverse and can exist as a both a commensal or a pathogen causing serious infections. How different strains regulate their responses to their surrounding environment is a key question and is illustrated below.
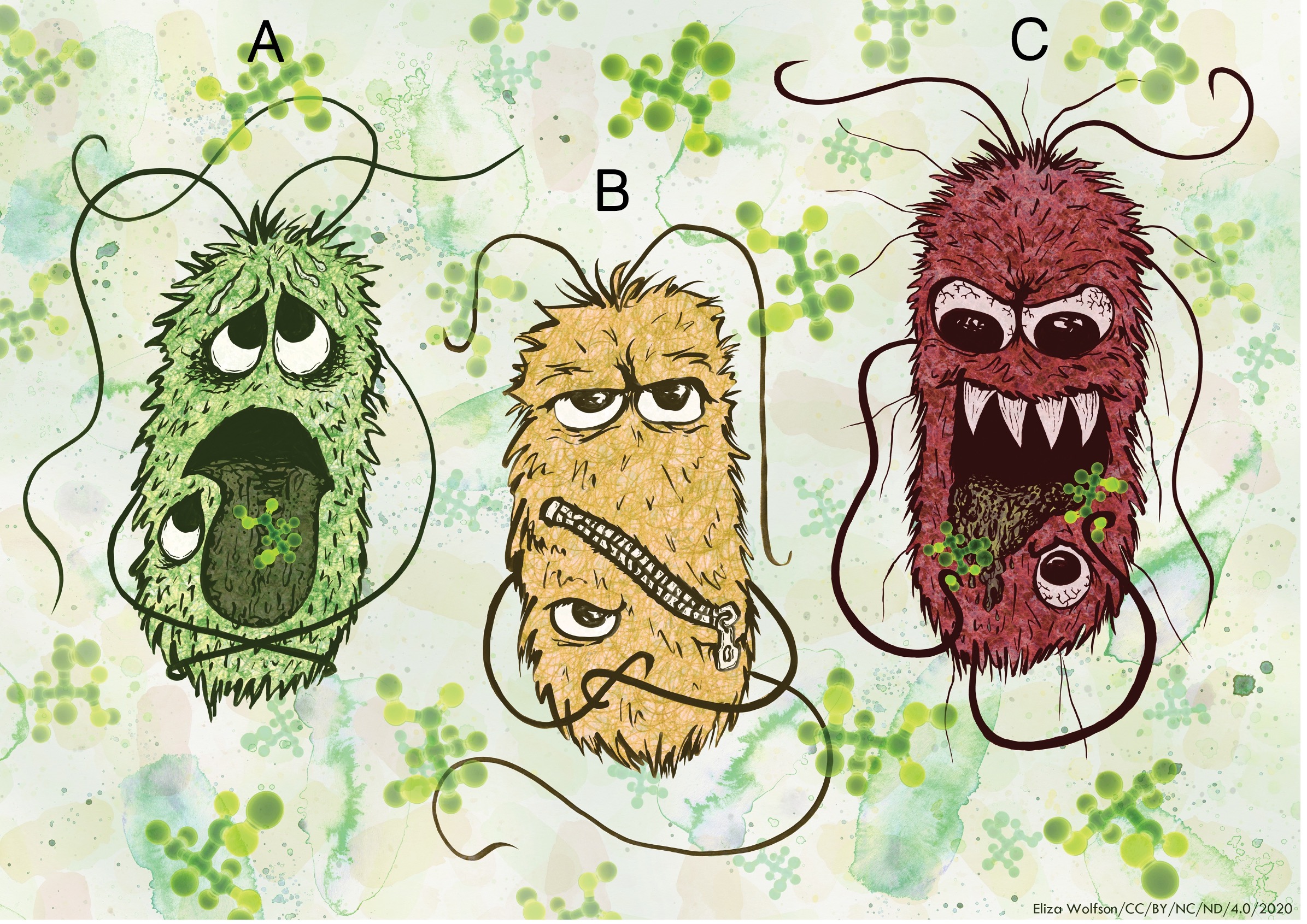
Figure 1: Image showing 3 different responses to the host metabolite D-serine. See O’Boyle et al., (2021).
As Escherichia coli is such a tractable organism, we utilize a wide variety of methods, ranging from microbiology and molecular biology, via biochemistry, to structural biology. We take full advantage our in house polyomics unit, a core facility at the University supporting transcriptomics, proteomics and metabolomics.
Grant funding from the BBSRC, Horizon EU and MRC provides support for our postdoctoral researchers: Rebecca McHugh, Clio Dritsa, David Mark & Emily Addington.
Currently the group hosts three PhD students: Emily Horsburgh, Sam Tucker and Charlotte Bryson covering a range of projects and techniques - see their individual pages for more details. Training and supporting early career scientists is a core philosophy for the group.
Past lab members have gone on to successful careers in many aspects of science and beyond. They form a key part of our science legacy and have contributed to over 90 papers and reviews. We are always keen to talk to motivated students or scientists at any stage of their career if they are keen to come to the lab for an experience, be it a summer placement, PhD project or personal Fellowship.
Featured Research
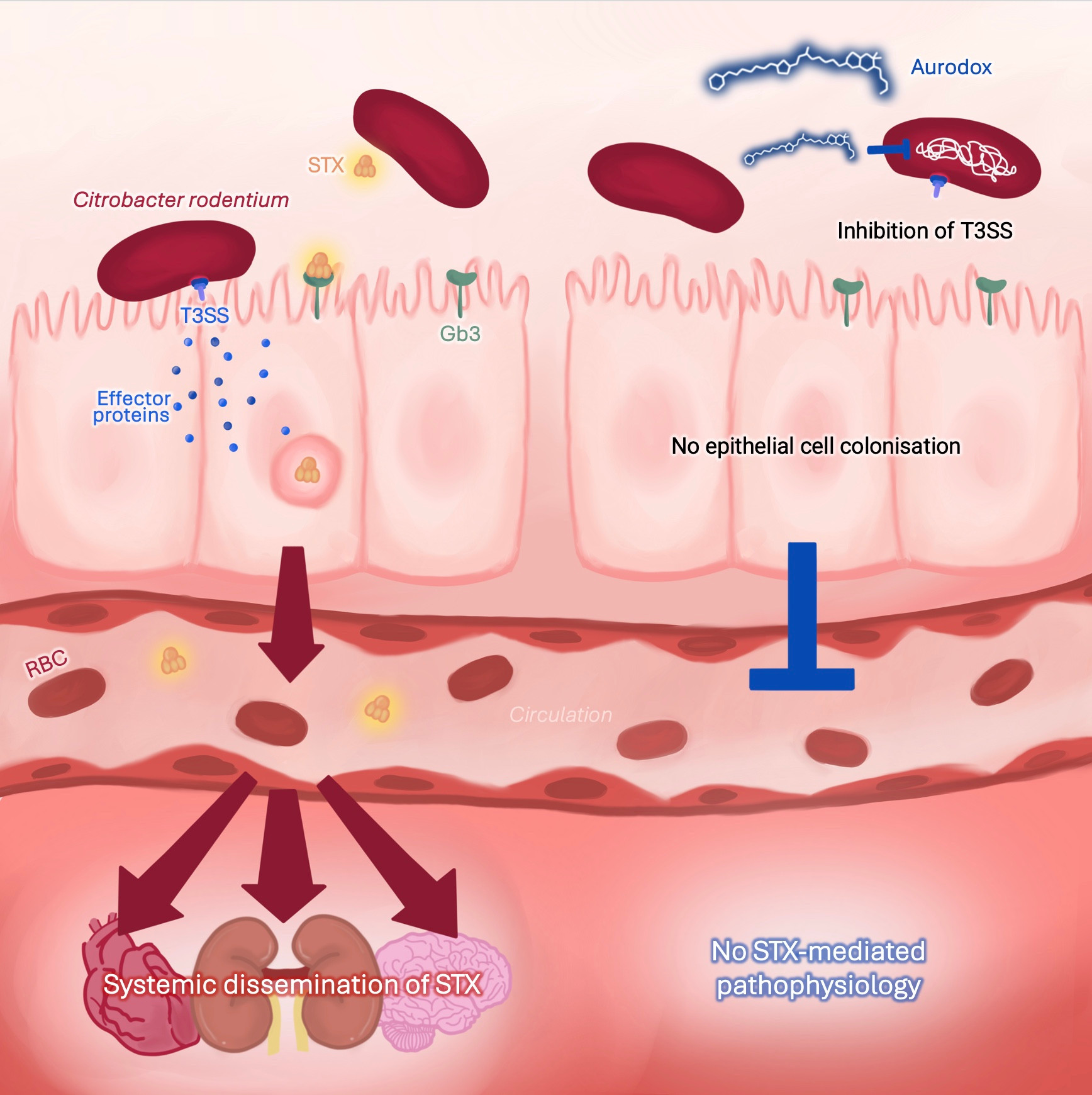
Evaluating the protective effects of Aurodox in a murine model of Shiga toxin-producing Escherichia coli
npj Antimicrobials and Resistance (2025)
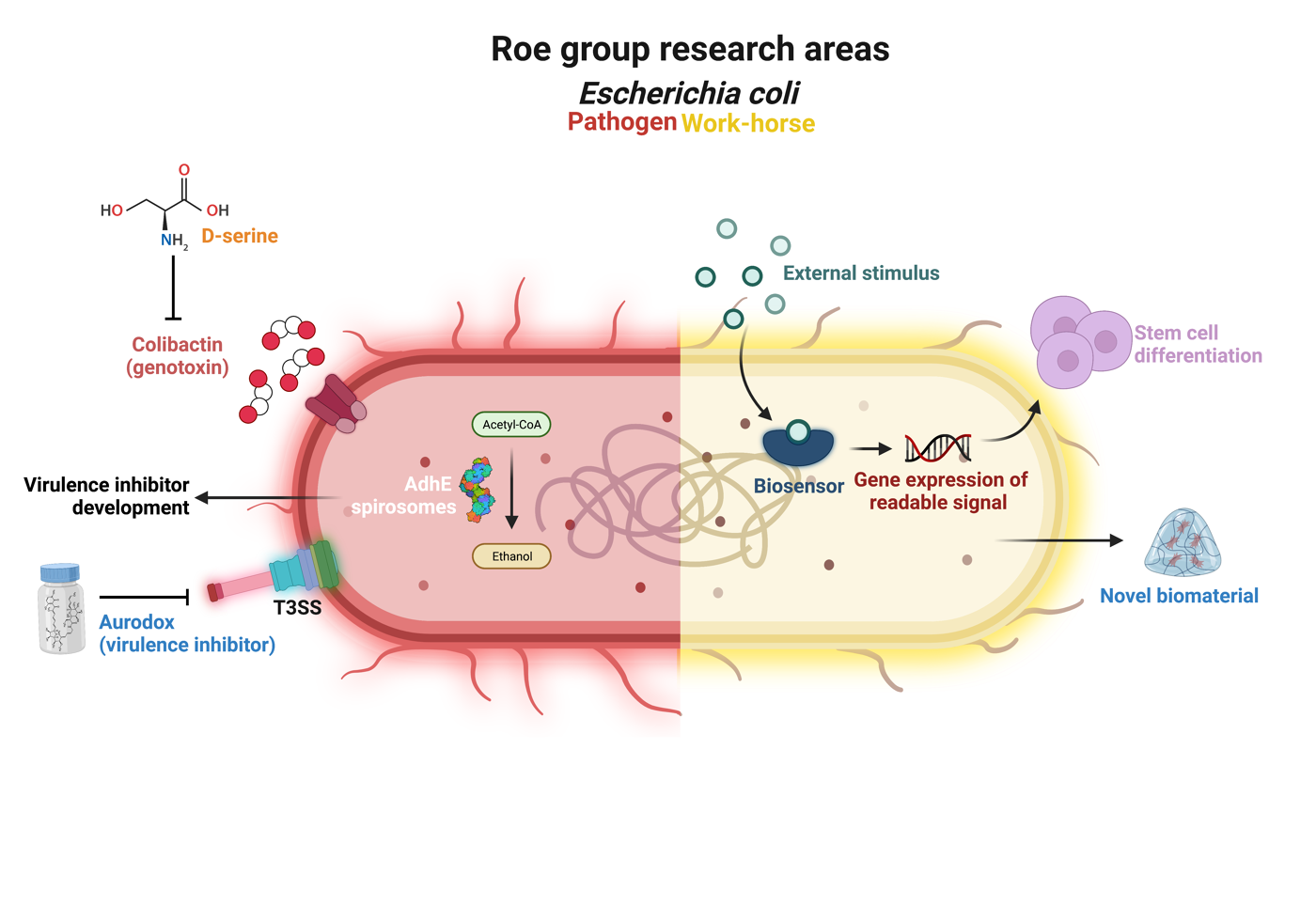
Research areas

Meet the team
Principal Investigator
Post-Doctoral Researchers
PhD Students
- Emily Horsburgh
- Sam Tucker
- Charlotte Bryson
Scientific Graphic Illustrator
Recent Publications
Niche specificity:
Wale, K. R., O’Boyle, N., McHugh, R. E., Serrano, E., Mark, D. R., Douce, G. R., ... & Roe, A. J. (2024). A master regulator of central carbon metabolism directly activates virulence gene expression in attaching and effacing pathogens. PLoS Pathogens, 20(10), e1012451, available: https://doi.org/10.1371/journal.ppat.1012451
O'Boyle, N., Douce, G.R., Farrell, G., Connolly, J., Roe, A.J., Schembri, M. and Rattray, N. (2022) 'Distinct ecological fitness factors coordinated by a conserved regulator during systemic bloodstream infection', Proceedings of the National Academy of Sciences of the United States of America, 120(1), e2212175120, available: http://dx.doi.org/10.1073/pnas.2212175120.
Connolly, J.P., Turner, N.C., Serrano, E., Rimbi, P.T., Browning, D.F., O’Boyle, N. and Roe, A.J. (2022) 'Control of resistance against bacteriophage killing by a metabolic regulator in meningitis-associated Escherichia coli', Proceedings of the National Academy of Sciences, 119(45), e2210299119, available: http://dx.doi.org/10.1073/pnas.2210299119.
Structure function:
Zambelloni, R., Beckham, K.S., Wu, H.-J., Elofsson, M., Marquez, R., Gabrielsen, M. and Roe, A.J. (2022) 'Crystal structures of WrbA, a spurious target of the salicylidene acylhydrazide inhibitors of type III secretion in Gram-negative pathogens, and verification of improved specificity of next-generation compounds', Microbiology, 168(7), 001211, available: http://dx.doi.org/10.1099/mic.0.001211.
Understanding and treating infections:
Mark, D. R., O’Boyle, N., Wale, K. R., Tucker, S. K., McHugh, R. E., & Roe, A. J. (2024). Aurodox inhibits type III secretion in multiple Gram-negative pathogens. Open Biology, 14(11), 240240, available: https://doi.org/10.1098/rsob.240240
Rimbi, P.T., O'Boyle, N., Douce, G.R., Pizza, M., Rosini, R. and Roe, A.J. (2024) 'Enhancing a multi-purpose artificial urine for culture and gene expression studies of uropathogenic Escherichia coli strains', Journal of Applied Microbiology, 135(4), lxae067, available: http://dx.doi.org/10.1093/jambio/lxae067.
McHugh, R.E., Munnoch, J.T., Roe, A.J. and Hoskisson, P.A. (2022) 'Genome sequence of the aurodox-producing bacterium Streptomyces goldiniensis ATCC 21386', Access Microbiol, 4(5), acmi000358, available: http://dx.doi.org/10.1099/acmi.0.000358.
McHugh, R.E., Munnoch, J.T., Braes, R.E., McKean, I.J., Giard, J., Taladriz-Sender, A., Peschke, F., Burley, G.A., Roe, A.J. and Hoskisson, P.A. (2022) 'Biosynthesis of aurodox, a Type III secretion system inhibitor from Streptomyces goldiniensis', Applied and Environmental Microbiology, 88(15), e00692-22, available: http://dx.doi.org/10.1128/aem.00692-22.
Reviews:
Tucker, S.K., McHugh, R.E. and Roe, A.J. (2024) 'One problem, multiple potential targets: Where are we now in the development of Shiga toxin inhibitors?', Cellular Signalling, 111253, available: http://dx.doi.org/10.1016/j.cellsig.2024.111253.
Addington, E., Sandalli, S. and Roe, A.J. (2024) 'Current understandings of colibactin regulation', Microbiology, 170(2), 001427, available: http://dx.doi.org/10.1099/mic.0.001427.


