Untargeted metabolomics highlight markers for PMI
Published: 26 October 2020
A project developed by PhD student Bogusia Pesko has utilised biomarker discovery technologies to establish a reliable, quantitative, and objective method for the estimation of time since death in human fatalities.
A project developed by Dr Bogusia Pesko has utilised biomarker discovery technologies to establish a reliable, quantitative, and objective method for the estimation of time since death in human fatalities.
Dr Pesko, a PhD student on the EPSRC/BBSRC Doctoral Training Centre in Proteomic Technologies coordinated by Dr Richard Burchmore, collaborated with an interdisciplinary team to investigate postmortem interval (PMI).
The findings, published online ahead of print by OMICS, provide proof of principle for an untargeted metabolomic approach, which could have potential applications in forensics as well as in the area of food safety.
In real life forensic investigations - and in television crime dramas - it is often important to estimate time of death.
However, this estimation is very subjective, particularly when a body has remained undiscovered for some days.
Even when available methods are combined, the margin of error can be 40 per cent - potentially leading to legal challenges.
There is thus a clear need for a reliable, quantitative, and objective method for the estimation of time since death.
This project exploited biomarker discovery technologies, developed to diagnose or predict disease, to reveal molecular markers that evolve predictably over time since death.
Findings from a controlled experiment with rat cadavers were mirrored in an inevitably less controlled experiment with human cadavers, providing proof of principle that this approach has potential to infer time since death.
Bogusia had previously completed an MSci in Forensic Toxicology, in which she used mass spectrometry to screen for drugs in forensic samples.
She joined the Burchmore Group with the goal of extending her skills in mass spectrometry and was able to access state-of-the-art facilities for metabolomics and proteomics, working both in the Institute of Infection, Immunity & Inflammation and Glasgow Polyomics.
With the support of a Lister/Bellahouston Travel Fellowship, Bogusia visited the Forensic Anthropology Center at Texas State University, where she collected samples from human cadavers donated for forensic research.
Markers for time since death have applications beyond forensics, particularly in the area of food safety and quality.
She is currently collaborating with colleagues in engineering to develop point-of-use devices to assess these markers in the field.
Bogusia said: "This project is a very unusual application of biomarker discovery technology. The interdisciplinary team has been very refreshing to work with.
"We hope the work will extend the application of this powerful technology beyond clinical research."

Postmortomics: The Potential of Untargeted Metabolomics to Highlight Markers for Time Since Death
- Bogumila K. Pesko, Stefan Weidt, Mark McLaughlin, Daniel J. Wescott, Hazel Torrance, Karl Burgess, and Richard Burchmore.
- OMICS: A Journal of Integrative Biology. Published Online: 13 Oct 2020, https://doi.org/10.1089/omi.2020.0084
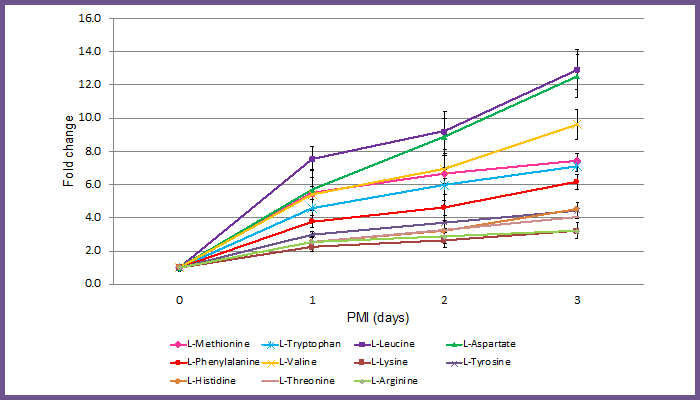
Image: Figure 2 - Fold change in signal for 11 amino acids in rat tissue with increasing PMI. Signal at each time point was normalized to time 0. Error bars indicate standard deviation between four samples. Two rats were analyzed per time point, from each hind limb, giving four replicate samples at each time point. PMI, postmortem interval.
First published: 26 October 2020

