African Trypanosomes
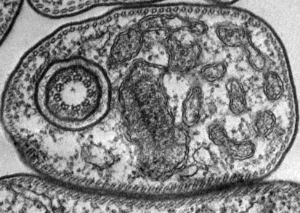
Trypanosomes are responsible for causing Human African Trypanosomiasis (HAT), also known as sleeping sickness. Trypanosomes are transmitted to humans through infected tsetse flies, which breed in warm and humid areas across sub-Saharan Africa. Tsetse flies also come into contact with cattle and wild animals which all act as reservoirs for the Trypanosoma parasites. The WHO estimates that 60 million people are at risk in 36 Sub-Saharan countries.The infection attacks the central nervous system causing severe neurological disorders, and is fatal without treatment. More than 95 percent of reported cases are caused by the parasite Trypanosoma brucei gambiense, which is found in western and central Africa. Animal African Trypanosomiasis (AAT) affects livestock in this region and is a huge impediment to economic development.
WCIP research focus
Trypanosome research in WCMP is long-standing and varied, based around understanding the molecular genetics, cell biology and biochemistry of the parasite and its interactions with the host. Specifically, we are exploring the following:
- Genome stability and transmission, including strategies for genome repair, replication and how these are exploited for antigenic variation.
- Parasite genetic exchange and mapping of virulence traits, including the ability of the parasite to withstand primate serum killing.
- The trypanosome life cycle including protein turnover and cellular remodelling.
- Resistance to anti-trypanosome drugs
- Parasite and host co-evolution
Next generation nucleic acid sequencing strategies are being applied, as well as mapping metabolic changes through metabolomic approaches. Potential drug targets that emerge from such work are being used for early stage drug discovery.
Key WCIP publications
Hall,J.P. et al. (2013) Mosaic VSGs and the scale of Trypanosoma brucei antigenic variation. PLoS. Pathog. 9, e1003502
Myburgh,E. et al. (2013) In vivo imaging of trypanosome-brain interactions and development of a rapid screening test for drugs against CNS stage trypanosomiasis. PLoS. Negl. Trop. Dis. 7, e2384
Trenaman,A. et al. (2013) Trypanosoma brucei BRCA2 acts in a life cycle-specific genome stability process and dictates BRC repeat number-dependent RAD51 subnuclear dynamics. Nucleic Acids Res. 41, 943-960
McLuskey,K. et al. (2012) Crystal structure of a Trypanosoma brucei metacaspase. Proc. Natl. Acad. Sci. U. S. A 109, 7469-7474
Tiengwe,C. et al. (2012) Genome-wide Analysis Reveals Extensive Functional Interaction between DNA Replication Initiation and Transcription in the Genome of Trypanosoma brucei. Cell Rep. 2, 185-197
Vincent,I.M. et al. (2012) Untargeted metabolomics reveals a lack of synergy between nifurtimox and eflornithine against Trypanosoma brucei. PLoS. Negl. Trop. Dis. 6, e1618
Kieft,R. et al. (2010) Mechanism of Trypanosoma brucei gambiense (group 1) resistance to human trypanosome lytic factor. Proc. Natl. Acad. Sci. U. S. A 107, 16137-16141
Morrison,L.J. et al. (2009) A major genetic locus in Trypanosoma brucei is a determinant of host pathology. PLoS. Negl. Trop. Dis. 3, e557
Vincent,I.M. et al. (2010) A molecular mechanism for eflornithine resistance in African trypanosomes. PLoS. Pathog. 6, e1001204

