Clinical Research Fellowships
Clinical Research Fellowships (CRFs) are an excellent opportunity for clinically qualified medical doctors to develop advanced training in a range of research methodologies relevant to modern biomedical research, and to consolidate their academic career prospects by undertaking a higher degree (normally a PhD).
The fellowships are intended for applicants with an interest in infectious diseases or clinical virology. In most cases applicants should be an internal medicine trainee (IMT), UK speciality trainee, or international equivalent level from any specialty. Applicants must be at pre-consultant level, GMC registered at the time of starting the post, and be able to gain out-of-programme (OOP) approval.
CVR Clinical Research Fellows are integrated into a wider cohort of postgraduate research students at the CVR. The training and support provided to these cohorts is described here.
For 2025, there are two opportunities to undertake a CRF with the CVR: CVR Clinical Research Fellowships and MRS-RCPSG Clinical PhD Studentship awards.

CVR Clinical Research Fellowships
Applications for August 2025 are now open. Closing date is 26 November 2024.
Project Selection
CRFs differ from the more usual CVR MRC PhD studentships in two ways. First, CRFs are funded for only three years; therefore, it is not possible to carry out rotations projects before starting your main thesis project. In order to complete a PhD project within a three-year timeframe and to allow a return to run-through training or timely applications for academic training posts, we strongly encourage applicants to consider potential project areas and to reach out to potential supervisors prior to interview. Secondly, CRFs usually start in August rather than October in order to fit in with clinical rotations. Despite the earlier start date, you are encouraged to take part in the ‘CVR Introduction to Virus Research’ course.
We encourage projects in all areas of CVR research (fundamental, translational, and clinical virology), from computational biology, to molecular biology, to epidemiology and beyond. Opportunities also exist for collaborative projects, including in tropical medicine and emerging diseases, with opportunities to spend time working with partner institutions overseas. Following interview, the successful applicant will be supported to finalise their choice of project before starting their fellowship.
An overview of the research groups in the CVR can be found in the booklet linked below, and examples of possible projects are given in the following section.
Introduction to Postgraduate Research at the CVR
Benefits
You will work in a world-leading virology research institution, alongside the UK’s largest grouping of virologists. Your training experience will centre on a 'hands on' research project in your chosen supervisor's laboratory, leading to the submission of a PhD thesis. This training will be supplemented with mentorship, career advice and the acquisition of transferable skills. Although research forms the major component of this post, clinical duties and teaching can be incorporated according to individual needs.
The salary for Clinical Research Fellows will be on the Clinical Academic pay scale, and the studentship includes all University fees as well as a generous allowance to cover consumables and travel (including international conference attendance).
Application Process
Timeline:
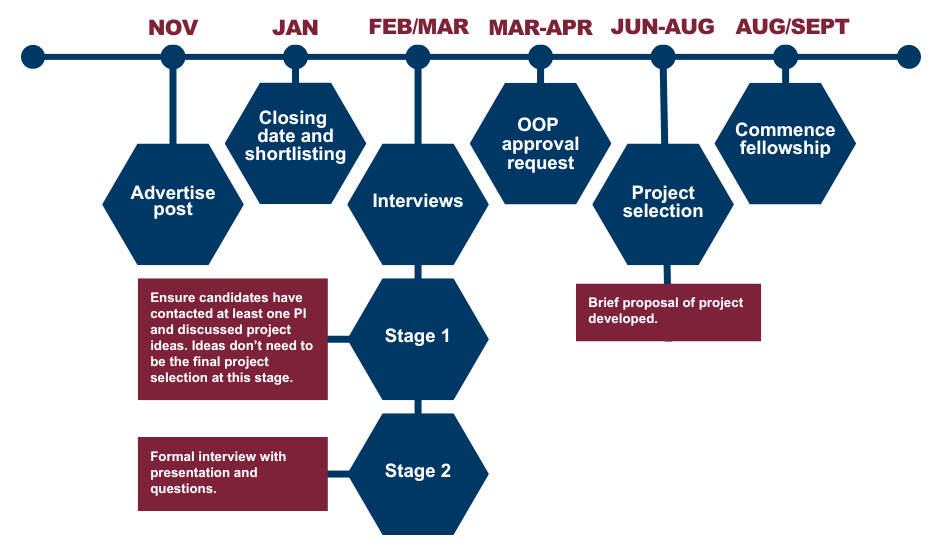
Shortlisted applicants will be invited to come to Glasgow to visit the CVR. Reasonable travel expenses will be reimbursed and accommodation made available as required.
Any questions about the programme or eligibility requirements should be directed to antonia.ho@glasgow.ac.uk.
MRS-RCPSG Clinical PhD Studentships
Medical Research Scotland (MRS) and the Royal College of Physicians and Surgeons of Glasgow (RCPSG) have launched their jointly funded 3-year MRS-RCPSG Clinical PhD Studentship awards.
The closing date for applications in 12 noon, Thursday 21 November 2024.
The awards are for a three year fully funded (at Home fee rate) full time Clinical PhD Studentship designed to provide a first-class Clinical PhD Studentship programme to research into any matter relating to human health that is clinical or patient focused.
Funded clinical students receive three years of clinical salary, up to £30,000 research expenses, £2,250 travel allowance and payment of university fees (home rate).
Please visit here for more information.

Clinical Project Fellows Stories
Find out from current and past fellows about their CRF projects.
Igor Starinskij

Start date: 2022
Project Title: Research tools for Erve virus, a neglected European nairovirus
Project Summary: Erve virus is a widespread, pathogenic, yet understudied virus that is associated with severe headache. Its basic structure and genomic sequence are known but there are few studies looking at the disease-causing potential of this agent.
The purpose of this project is to set up an array of tools to allow further investigation into Erve virus. First, detection methods will be developed: antibody test, RT-qPCR, pseudoneutralisation assay. These will then be employed on surveillance samples of UK wildlife and humans with undiagnosed disease in an attempt to identify the presence of Erve virus in the UK. These assays will be essential for anyone undertaking future investigations into this pathogen in the future.
Second, laboratory research tools, such as reverse genetics, will also be developed and used for fundamental virus characterisation. These methods will permit the wider scientific community to study Erve and related viruses further.
Project Experience: Working in a purely academic setting is both rewarding and challenging. It is fantastic to have the freedom to pursue novel ideas as well as an autonomy over your workload and time. Nonetheless, exploring something new can be frustrating, with some ventures failing to yield results. This endeavour requires tremendous initiative and an ability to be self-critical. Something I have not yet mastered is making sure the experiences of today will be useful in future academic work. I would therefore advise to have a career plan which, with inevitable amendments, of course, will maximise your success after the PhD is completed.
James Sheperd

Start date: 2017
Project Title: Detection and characterisation of emerging rhabdoviruses in Uganda
Project Summary: My project focused on detection of viruses spilling over into human populations in Uganda using a technique called metagenomic next generation sequencing. This involved recruitment of a cohort of patients presenting with acute febrile illness in Northern Uganda. In addition, I used serological techniques to determine the population exposure to novel viruses in Uganda and also conducted sampling of various potential zoonotic reservoir species such as bats and small mammals.
Project Experience: I began my project with a very clinical focus and a plan to recruit a cohort of patients with acute febrile illness in Uganda. By the end I found I had developed skills in project management, tissue culture, molecular biology, pathogen ecology, next generation sequencing and bioinformatics. Working at the CVR allowed me to interact with experts in broad range of disciplines and use these new tools to ask clinically relevant questions. My PhD project was initially quite loosely defined, and much of the early process involved developing the ethics proposals, study design and research plans. The first few years were a steep learning curve but by the end of my fellowship I had developed the skills and understanding of the field to develop scientific questions and hypotheses. My advice would be to not box yourself in early on – for example even if you think you are not a “wet lab person” it's worth giving it a try and pushing beyond your comfort zone. You may find you like it and developing a broad skillset and experience of different disciplines is key to understanding what is possible in clinical research. As an example – my understanding of the principles of next generation sequencing and viral genomics proved extremely useful in response to the COVID-19 pandemic where I was able to pivot from my PhD project to focus on the research response to SARS-CoV-2.
John Kelly

Start date: 2024
Project Title: Arboviral Infection in Malawi: Serology and Epidemiology
Project Summary:
The global burden of mosquito-borne viruses like Dengue, Zika, and Chikungunya has rapidly grown in recent decades. However, their impact in Africa is not well understood. These infections are not captured by existing surveillance systems in Malawi, and they are often overlooked as causes of fever in malaria-endemic contexts. Historically, flavivirus serology has been complicated by high degrees of cross-reactivity, limiting the performance of assays in co-endemic settings.
The first aim of this project is to develop sensitive and specific NS1 protein-based ELISAs for the detection Dengue and Zika IgG. These will be used alongside pseudotype neutralisation assays for exposure to Chikungunya and O’Nyong-nyong, to evaluate sera from a serial surveillance study at two urban and two rural sites in Malawi. The overall aim is to characterise the seroprevalence and epidemiological risk factors for exposure to mosquito-borne viral infections across Malawi, through analysis of linked clinical and demographic data. Development of affordable, reliable assays will make it possible to describe the seroepidemiology of these infections in Malawi, and build capacity for surveillance. Identify risk factors for infection will guide further research to help direct public health interventions such as vaccination.
Project Experience:
Having only recently started, I have limited experience in the trials and tribulations of a PhD. However, so far I have enjoyed having the time to really delve into a research project – rather than the snatched moments after hours during clinical work. My new colleagues have been supportive and welcoming, helping me get to grips with the lab. The change of pace from clinical work has been welcome, but also difficult to adjust to when I have been excited to get my research project up and running.
I know that there will be challenges, and the prospect of developing novel serological assays does feel a little daunting. I am also aware that PhD students (and especially clinicians) can spend much of the early part of projects just learning how to be a scientist, rather than generating lots of data. I’m excited to learn, but also wary of the frustration this can bring, particularly as I will need to develop skills in both wet and dry lab disciplines. The increased flexibility compared to NHS work is a major advantage, accommodating other responsibilities like childcare and on-call work, and some working from home.
Kathy Li
Project Summary: The betaherpesvirus human cytomegalovirus (HCMV) is a ubiquitous viral pathogen. It is the most common cause of congenital infection in infants and of opportunistic infections in immunocompromised patients worldwide. The large double-stranded DNA genome of HCMV (236 kb) contains several genes that exhibit a high degree of variation among strains within an otherwise highly conserved sequence. These hypervariable genes encode immune escape, tropism or regulatory factors that may affect virulence. Variation arising from these genes and from an evolutionary history of recombination between strains has been hypothesised to be linked to disease severity. To investigate this, the HCMV genome has been scrutinised in detail over the years using a variety of molecular techniques, most looking only at one or a few of these genes at a time. The advent of high-throughput sequencing (HTS) technology 20 years ago has enabled more in-depth whole-genome analyses. My study extends this field by using both HTS and the more recently developed long-read nanopore technology to determine HCMV genome sequences directly from clinical samples.
Firstly, I used an Illumina HTS pipeline to sequence HCMV strains directly from formalin-fixed, paraffin-embedded (FFPE) tissues. FFPE samples are a valuable repository for the study of relatively rare diseases, such as congenital HCMV (cCMV). Next, I developed a pipeline utilising the single-molecule, long-read sequencer from Oxford Nanopore Technologies (ONT) to sequence HCMV initially from high-titre cell-cultured laboratory strains and then from clinical samples with high HCMV loads. Finally, I utilised a direct RNA sequencing protocol with the ONT sequencer to characterise novel HCMV transcripts produced during infection in cell culture, demonstrating the existence of transcript isoforms with multiple splice sites. Overall, my findings demonstrate how advanced sequencing technologies can be used to characterise the genome and transcriptome of a large DNA virus, and will facilitate future studies on HCMV prognostic factors, novel antiviral targets and vaccine development.
As a medical virologist working in a diagnostic laboratory, my PhD has helped bolster my knowledge and experience in using these advanced sequencing techniques both in the practical laboratory aspect as well as in data-processing and bioinformatics; this has put me in good stead as this technology is implemented more widely.
Leah Owen

Start date: 2023
Project Title: Understanding zoonotic spillover and epidemiology of orthonairoviruses, focusing on Crimean Congo Haemorraghic Fever (CCHFV) in Uganda.
Project Summary: My PhD focuses on better understanding the epidemiology and genetic diversity of CCHFV, primarily in Uganda. CCHFV is a widely distributed tick-borne virus causing a spectrum of disease, ranging from asymptomatic infection to a severe haemorrhagic fever with a very high case fatality rate. The epidemiology and burden of infection are still poorly understood, particularly in African settings and it likely represents an underdiagnosed cause of febrile illness. CCHFV strains demonstrate much genetic diversity between geographical regions, with the drivers and implications of these molecular changes not yet determined.
The three main objectives of my work are firstly, to develop a publicly accessible database of global CCHFV genetic sequences and relevant metadata, and use this to characterise global genetic diversity in CCHFV, investigating potential drivers and effects of genetic variation between different virus strains. Secondly, to determine prevalence and diversity of nairoviruses (including CCHFV) within tick populations across distinct ecological regions of Uganda and to compare this with nairovirus exposure in human populations in these regions. Finally, to determine the annual incidence of human CCHFV infection across six sites in Uganda through a longitudinal seroepidemiological study.
Project Experience: Moving from clinical work to a PhD has been rewarding but challenging, particularly as it also involved moving to a new city (and country!). A PhD project tends to be far less structured day-to-day than clinical work, which takes a bit of getting used to and requires good self-management skills. However, this also allows greater flexibility and autonomy in your working pattern which has been beneficial when juggling work and caring commitments. Moving from working in a large busy hospital team to an academic environment with smaller teams and more solo working also takes some adjustment, however my project involves a lot of collaborative work with colleagues from many different disciplines both in Glasgow and Uganda which is one of the most rewarding parts of my PhD.
My project includes wet and dry laboratory and field work and trying to learn many new skills simultaneously has been difficult and progress can feel slow. It is challenging feeling like a ‘beginner’ again after many years of clinical training. However, many of the problem solving and organisational skills learnt through clinical training have been very helpful during my PhD. To make the most of your PhD I think it is important to remain flexible and both adapt to disappointments for example when ideas do not pan out or field work is delayed and take opportunities when your work leads you in new directions. Oh, and remember that everything always takes much longer than you think it will!
Natasha Jesudason

Start date: 2018
Project Title: Inherited chromosomally integrated human herpesvirus 6: evolution and clinical significance
Project Summary: Inherited chromosomally integrated HHV 6 (iciHHV-6) occurs in approximately 1% of the global population and is the result of single integration events that have occurred thousands of years ago. To date, little is understood about the prevalence of iciHHV-6 and potential disease associations.
This project has three main aims: firstly, to describe the prevalence and characterise the integration sites of ancient lineages of iciHHV-6 in large cohort studies of isolated populations in Europe using a range of laboratory and data-mining methods. Secondly, the potential clinical impact of iciHHV-6 is being explored by analysis of linked electronic health record data from the Generation Scotland cohort in whom iciHHV-6 has been previously screened by the Jarrett group. Thirdly, clinical assays are under development to aid in the differentiation of iciHHV-6 from exogenous HHV-6 which is a frequent diagnostic dilemma faced by clinical virologists in the management of transplant recipients.
Project Experience: Embarking on a PhD, especially a lab-based project requires a significant change in approach compared to work as a clinical trainee. I think it is important to have a clear idea of the research skills and the professional goals you hope to achieve prior to starting your project and communicate these with your supervisors so that together you can direct your PhD research project in a way that allows you to meet these over your study period. One of the biggest adjustments for me has been overcoming the frustration of experiments not working and planning for potential ‘setbacks’. Goals are met over a much longer time period than is usual in clinical work, however, seeing particular aspects of my project near completion has been incredibly fulfilling. My PhD journey has been slightly different to that of most clinical research fellows and one of the benefits for me has been the flexibility with which I can manage my time.
Rajiv Shah

Start date: 2019
Project Title: Hepatitis C Virus Diversity in Sub-Saharan Africa and implications for treatment with Direct Acting Antivirals
Project Summary:
HCV is a leading cause of chronic liver disease worldwide with an estimated 51 million suffering with chronic HCV infection and approximately 1.5 million new infections occurring each year. The WHO has set ambitious goals to eliminate viral hepatitis by 2030. A key strategy behind these goals is the discovery of Direct Acting Antivirals (DAAs), which have revolutionised the treatment of HCV infection. However most clinical trials and real world DAA treatment programs have been conducted in high income countries where the predominant circulating -genotypes are epidemic lineages. However, in Sub-Saharan Africa endemic lineages are more common and less well characterised, with growing evidence that certain sub-genotypes are more challenging to treat, for example 1l and 4r. The aims of this thesis were to explore the diversity of HCV using datasets from Uganda and Benin. A sub-genomic replicon system was used to study the effects of DAAs on HCV replication of previously uncharacterised subtypes in-vitro. Furthermore, the prevalence of HCV and associated liver disease from people who inject drugs (PWID) in Coastal Kenya was assessed and HCV genetic data was used to explore PWID networks and estimate the origin of HCV in this community.
Project Experience:
Doing a PhD full-time was a big change to doing daily clinical work, which is what I was used to for many years. The first year was challenging in a number of ways; 1) time management and structure is entirely self-governed, unlike clinical work, therefore this took some time to adjust to, 2) things don’t always go to plan and dealing with this disappointment can be difficult, 3) doing dry and wet laboratory work was learning a new discipline entirely and was overwhelming. However, as the data started to make sense and laboratory work became successful, the project came together, and this was very rewarding. The opportunity to publish some papers was extremely satisfying.
The biggest differences from clinical work were the lack of a structured schedule and time management. However, I came to understand and appreciate this through my PhD. Finally, I think it’s useful to have an open mind when starting a PhD. Setbacks are common, and being flexible to accept other opportunities that may not be part of the original plan is an advantage.
Examples of Projects for Clinical Research Fellows
Research at the CVR spans investigations from the molecular, structural and cellular levels through to the individual host and affected population, allowing the integration of molecular and structural virology, cell biology, pathogenesis, epidemiology and mathematical modelling into your PhD project.
Clinical PhD Fellows are strongly encouraged to think about projects in advance of interview and to make contact with potential supervisors.
Examples of possible projects include (but are by no means limited to) the following:
Dr Chris Illingworth is interested in better understanding and preventing nosocomial transmission of viral illness.
The SARS-CoV-2 pandemic highlighted the importance of nosocomial transmission as a key problem in hospitals, with a study at one hospital estimating that as many as 15% of cases in the hospital were caused by this route. Recent work in viral genomics in the Illingworth group has identified new methods in virus genomics as being of use for understanding outbreaks in retrospect, and tracking outbreaks as they occur in real time. We carried out a provisional analysis of the role of PPE in nosocomial transmission, studying the consequences of FFP3 mask usage in a hospital in Cambridge. Via collaboration we have also explored the experience by health care workers of receiving real-time genomic data in real time. Genomic datasets generated during the pandemic provide a rich resource for retrospective analysis.
The aim of this project will be to explore a broad range of approaches combining genomic and other data to better understand and prevent nosocomial transmission. Potential avenues of exploration within the project include i) The development and use of simple methods in bioinformatics to better understand evolutionary relationships between viral sequences collected during an outbreak; ii) The use of more advanced methods for network reconstruction to elucidate patterns of viral transmission; iii) The exploration of methods from computational fluid dynamics to understand the role of the hospital environment in promoting or preventing transmission events; iv) Approaches to optimising the communication of genomic data to healthcare workers.
This project will provide training and experience in viral genomics, in bioinformatics, and in the use of quantitative methods for studying data from clinical settings.
Dr Chris Boutell and Professor Alfredo Castello are working to define the host-cell immunological response and barriers to MPOX infection.
MPOX (formerly known as monkeypox) is a disease caused by the MPOX virus (MPXV), a zoonotic pathogen transmitted from rodents to humans. Since January 2022, the WHO has reported a significant global rise in MPOX, totalling 86,309 confirmed cases and 107 deaths from 110 Member States across six WHO regions (https://worldhealthorg.shinyapps.io/mpx_global/). A high proportion of these cases have been reported from previously non-endemic regions, including Europe and the Americas (Thornhill et la., NEJM, 2022; Lum et al., Nat Rev Immunol, 2022). While MPOX is typically self-resolving, MPXV infection of the young or immunocompromised can lead to severe disease and increased likelihood of fatality. Human-to-human transmission occurs through contact with infected skin lesions, bodily fluids, and large respiratory droplets. Although smallpox vaccination and antiviral therapy (tecovirimat) can reduce MPOX replication and transmission, these countermeasures are not widely available or accessible in most countries. Thus, there is concern that MPXV may establish an endemic foot hold around the globe leading to an increase in MPOX morbidity and mortality within vulnerable groups. This MRC-funded Clinical Fellow (CF) project will investigate the host-cell tropism and immunological barriers to MPXV infection employing a range of two- and three-dimensional tissue culture model systems. Utilising a combination of genomic (RNA-seq) and proteomic technologies, this project will investigate whether strain dependent MPXV adaptation influences the kinetics of MPXV replication, activation of innate immune pathways, and sensitivity to immune suppression by the interferon response. This project will actively collaborate with our internal and external MPOX consortium partners to rapidly disseminate its findings in response to an ongoing outbreak.
This project will provide extensive training in infectious disease research across multiple technological platforms and research disciplines. The CF will actively participate in an existing collaboration between the Boutell group (CVR), other UKRI MPXV consortium members, and members of the CVR from Tissues to Molecules; Host-Cell Response to Infection programme to rapidly disseminate its research findings. This project will enable access to specialist knowledge and equipment not commonly found within an individual host institution or laboratory, for example BCL3 containment facilities, high-resolution light microscopy imaging, and bioinformatic support/training (MRC-UoG CVR). Outputs from this project will aid in the identification and characterisation of immune barriers and antiviral compounds that may restrict the replication or future re-emergence of zoonotic pathogens.
Prof Ed Hutchinson is interested in how respiratory viruses spread within hosts and between species
The Hutchinson group has a particular focus on influenza viruses, which are major pathogens of humans and livestock and which have an unusual ability to cross between species and cause pandemics. We try to understand these viruses at the scale of their cell and molecular biology, and often collaborate with other groups within and beyond the CVR to ask wider questions about how the virus works, at levels from protein structures to organ systems. Currently, multiple projects in the group are looking at how influenza virus infections establish themselves inside the host organism, and at the barriers preventing influenza viruses from one host species infecting another. If you’d be interested in devising a project in this or a similar area, please contact Ed Hutchinson to discuss this informally.
Prof Emma Thomson focuses on the use of NGS and functional assays to identify and investigate viruses that present a risk to human health in the UK and Uganda
Project 1: The orthonairoviruses CCHFV, Dugbe virus and Nairobi Sheep Disease virus are common in Uganda and other surrounding countries. A novel virus in the same family called Macira virus has also been identified recently, from tick collections in Uganda, by the Thomson group. The aim of this project is to investigate the role of cross-specificity in serological responses to these viruses by carrying out neutralisation assays using a non-infectious VLP system. Further, we will carry out a survey of ticks in Uganda to identify these viruses and to identify factors that may affect their distribution across the country.
Project 2: The group have recently identified a virus called adeno-associated virus 2 (AAV2) in children with otherwise unexplained hepatitis. The aim of project 2 is to investigate the immune response to AAV2 in children affected by this condition using ELISpot, flow cytometry, peptide binding assays and mass spectrometry.
Dr Louisa Pollock, Prof Brian Willett, Prof Antonia Ho are working to understand maternal and infant predictors of RSV vaccine effectiveness
RSV is the most common cause of respiratory infection in children worldwide, with the majority of severe disease in infants.1 New RSV immunisation strategies should reduce this burden of disease.2–4 The UK has introduced RSV vaccination in pregnant women >28 weeks’ gestation and adults >75 years. Several other countries have instead opted to introduce neonatal immunisation with nirsevimab.
The primary mechanism of protection in maternal immunisation is placental transfer of IgG antibody. Infants born prematurely have reduced time for antibody transfer. Maternal factors associated with placental insufficiency may lead to reduced antibody transfer and preterm birth. There are well-established data on the effect of maternal morbidities such as HIV and malaria, but limited data to determine whether other conditions associated with placental insufficiency, including smoking, obesity and diabetes have a similar effect. Rates of maternal multi-morbidities, obesity, and diabetes are very high and increasing in Scotland. Many of the risk factors for reduced placental antibody transfer are also known risk factors for severe respiratory infection in infants, including deprivation as an over-arching risk. This may mean that a maternal vaccine programme may be more likely to fail those most in need of protection.
We aim to determine the relationship between maternal risk factors for placental insufficiency, placental antibody transfer, and risk of RSV in infancy. As part of a larger programme of work we plan to undertake an active surveillance birth cohort, recruiting representative mother-infant pairs in NHS Greater Glasgow and Clyde, collecting paired mother-infant blood and placenta samples at birth with active follow up determine the relationship between maternal and infant risk factors, vaccine timing, placental antibody transfer and risk of RSV infection in the first 3 months of life.
Active surveillance approaches will determine vaccine effectiveness against mild RSV disease in infancy, informing modelling of vaccine-era community transmission. Identifying modifiable maternal risk factors for reduced maternal vaccine effectiveness could inform the focus of future public health strategies to improve maternal and infant outcomes. Finally identifying high-risk infants could inform new dual-approach RSV immunization strategies, highlighting infants who may benefit from neonatal immunisation in addition to maternal vaccination.
This project would offer opportunities to develop experience in perinatal immunology, clinical epidemiology and maternal determinants of infant health.
Prof Pablo Murcia, Ross Langley and Rory Gunson and interested in why infants and young adults have more viral coinfections than older children and adults
Prof Pablo Murcia, in collaboration with Ross Langley (QEUH) and Rory Gunson (West of Scotland Specialist Virology Centre), is interested in why infants and young adults have more viral coinfections than older children and adults.
Background. Respiratory viral infections are responsible for major disease burden. Different viruses can cause respiratory infections, including influenza viruses (IAV), coronaviruses (CoVs), respiratory syncytial virus (RSV), human rhinovirus (HRV), human metapneumovirus (hMPV) and parainfluenza viruses (PiVs), to name but a few. We have shown that viral coinfections represent ~10% of viral respiratory infections and that most of them occur in children under five years of age.
The interferon (IFN)-mediated innate immune response is the first cellular response against viral infections, and we and others showed that this response plays a key role in viral interference within the respiratory trac. For example, HRV, IAV and RSV trigger an IFN response that blocks SARS-CoV-2 replication. In contrast, RSV replication is minimally affected by IAV. This suggests that IFN-mediated virus interactions are specific and depend on i) the host's ability to mount an innate immune response; ii) the breadth and potency of the innate immune response triggered by each virus; and iii) the susceptibility of a given virus to the IFN response triggered by other viruses.
It is unclear why viral coinfections are more common in young children than in other age groups, and why certain viruses are more common than others in coinfections. Possible explanations are that very young children (0-3 years old) mount weaker innate immune responses than older children, and that certain viruses modulate the innate immune response in a way that facilitates viral coinfections. This project aims to address those two knowledge gaps by comparting the response to RSV infection and RSV/HRV coinfection in patients of different age groups. To this end we will generate transcriptomes from nasopharyngeal swabs collected from patients of different ages with a positive qPCR diagnosis for RSV, HRV, as well as coinfected with both viruses.
Results from this work will provide important insights on the impact of age on the response to virus infection and coinfection. Characterising the host response to viruses in patients is critical to identify markers associated with susceptibility to viral infection and/or altered disease outcomes.
Prof. Brian Willett and Prof. Margaret Hosie are interested in the development & application of techniques for analysis of humoral immune responses to viruses
Prof. Brian Willett & Prof. Margaret Hosie, in collaboration with Dr. Louisa Pollock (Queen Elizabeth University Hospital, Glasgow) & Dr. Helen Parry (Institute of Immunology and Immunotherapy, University of Birmingham) are interested in the development and application of novel techniques for the analysis of humoral immune responses to viral pathogens
Public Health Scotland (PHS) conducts community acute respiratory infection (CARI) surveillance to monitor respiratory infection across the country, to understand the burden of disease within the community and to estimate its impact on well-being. Samples are collected from representative sentinel practices and are screened for respiratory pathogens, including influenza, adenovirus, human metapneumovirus, Mycoplasma pneumoniae, parainfluenza, respiratory syncytial virus, rhinovirus, seasonal coronaviruses and SARS-CoV-2. A complementary scheme monitors severe acute respiratory infection (SARI) in secondary care. Together, these surveillance schemes enable PHS to contribute data to both the Scottish Government to formulate and refine healthcare policy, and to UK and international bodies such as JCVI and WHO to inform strategies for vaccination against influenza. At present, there are few national programmes investigating either serological evidence of pathogen exposure, or levels of immunity to infection in the general population. Healthcare strategies are focussed on achieving the levels of vaccine coverage that are predicted to confer sufficient immunity in the community to reduce onward transmission and prevent disease. For example, vaccine responses to measles, mumps and rubella are seldom measured following administration of the MMR vaccine; health boards simply aim to meet targets for vaccine coverage (>95% MMR coverage will prevent the spread of a measles virus with an R0 = 18 in a community).
This project aims to generate data with which healthcare policy can be better informed. The project will develop high-throughput assay systems with which functional immune responses can be measured rapidly and reliably. For example, as an adjunct to monitoring the current contribution of seasonal coronaviruses to acute respiratory infections, the project could assess the current level of immunity to seasonal coronaviruses (NL63, 229E, HKU1 & OC43), and other viral infections in the general population, identifying the demographic factors that influence immunity. For measles, current vaccines (eg M-M-RVaxPro and Priorix) are based on the genotype A Edmonston strain of measles virus. Circulating genotypes such as B3, C2, D8 and H1 might not be neutralised as effectively by responses elicited by current vaccination regimes. Moreover, immunity levels may differ significantly between children, adults, immunosuppressed and elderly individuals. Simple modifications to the number of doses administered, and the timing of doses could boost immunity sufficiently to confer longer-lasting immunity against all circulating genotypes.
This project will provide an opportunity to develop novel techniques with which antiviral immune responses can be measured. These techniques will then be applied to the analyses of sera from representative patient cohorts to assess levels of immunity in the general population.



