Research Interests
Research Interests
Epstein-Barr virus
One area of research in the laboratory concerns the analysis of the cancer-associated virus, Epstein-Barr virus (EBV). EBV is a human Herpesvirus that is causal in the development of several forms of human cancer, including Burkitt’s lymphoma, nasopharyngeal carcinoma, Hodgkin’s disease and others.
The virus leads to a life long infection, avoiding eradication by the immune system and has evolved intriguing tricks to do this.
We have developed transgenic models expressing key viral genes in order to investigate their mechanism of action in disease processes and to ask: how do they work to disturb the normal function of the cell, how does this lead to an abnormal cell state, how does this proceed to tumour formation and how does the virus avoid immune eradication? A critical viral gene is the EBV nuclear antigen-1 (EBNA1).
It is expressed in all viral associated tumour cells and is essential for viral DNA propagation, as well as enabling cell survival; a property which is likely involved in the development of cancers associated with this virus. EBNA1 represents a prime target for therapeutic design. To facilitate this, we have used in silico methods to develop a full structure for the protein.
From this we can see how the two monomers of EBNA1 “lock” together to form a stable dimer and how the homodimeric protein binds to DNA (Hussain et al (2014) Virus Genes 49: 358-372).
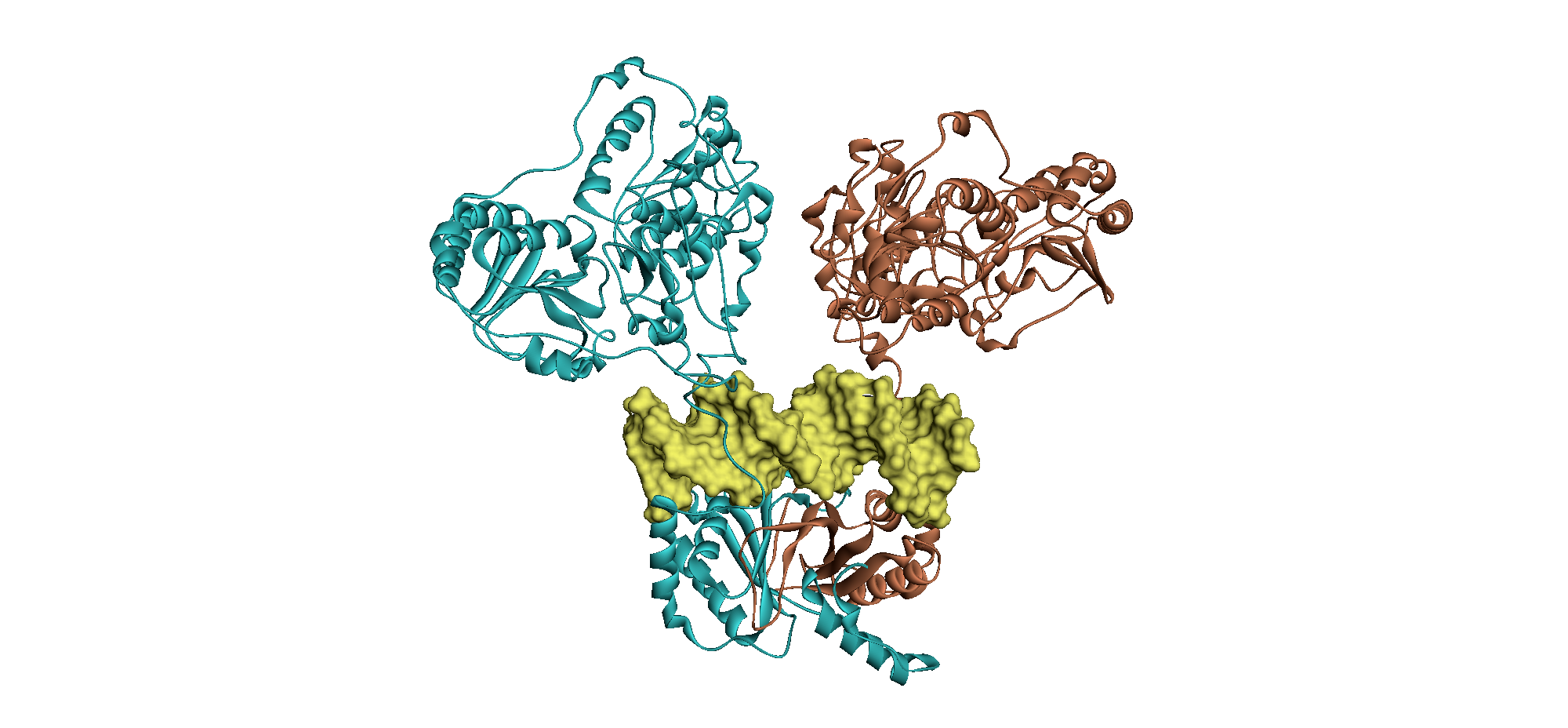
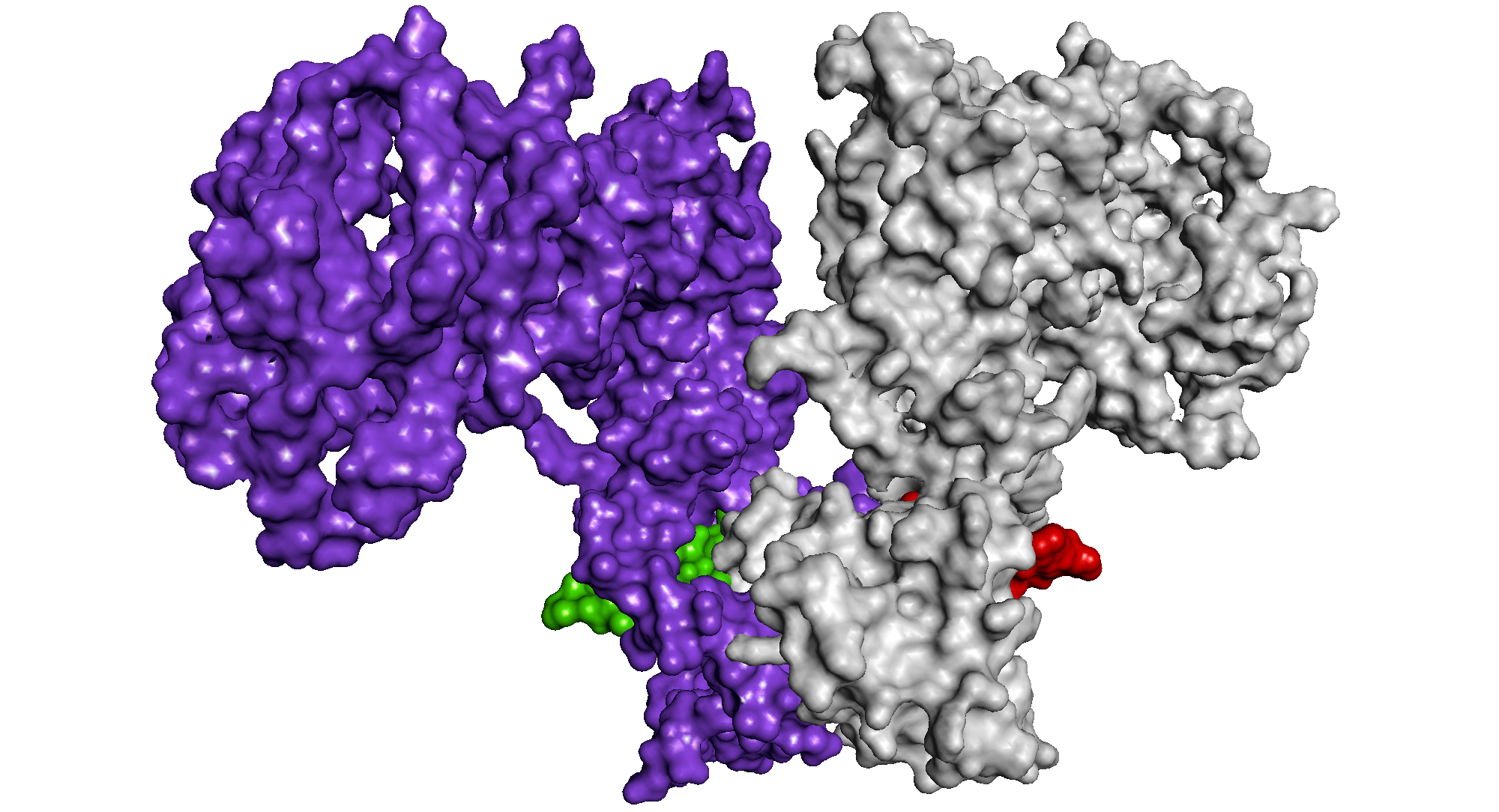
From: Hussain et al (2014) Virus Genes 49: 358-372. These images show an EBNA1 dimer in ribbon format (above: one monomer in cyan, the other brown), modelled with bound DNA (yellow) and an EBNA1 dimer in surface format (below: one monomer purple and red, the other silver and green), modelled to show the interlocking structure.
Another key gene of EBV encodes the latent membrane protein 1 (LMP1). This protein manifests many of the properties of a classic oncoprotein. It acts as a constitutively active signaling molecule and activates multiple signal transduction cascades leading to cell proliferation, cell survival and angiogenesis. We have found that it also induces inflammation. Hannigan et al (2011) Molecular Cancer 10:11 PMID: 21291541 DOI: 10.1186/1476-4598-10-11
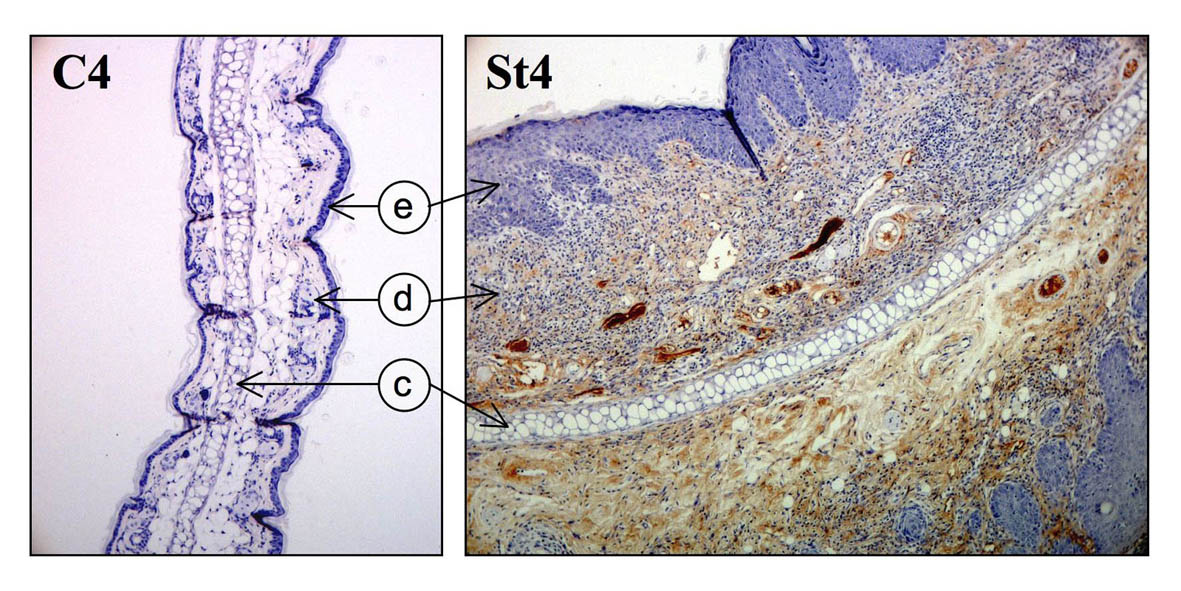
From: Hannigan et al (2011) Molecular Cancer 10:11 PMID: 21291541 DOI: 10.1186/1476-4598-10-11 (e=epidermis, d=dermis, c=cartilage). This image shows the extensive deposition of immunoglobulin G (IgG) as part of the inflammatory response, seen as a brown stain in the dermis of a skin section expressing LMP1 (St4) compared to a control (C4).
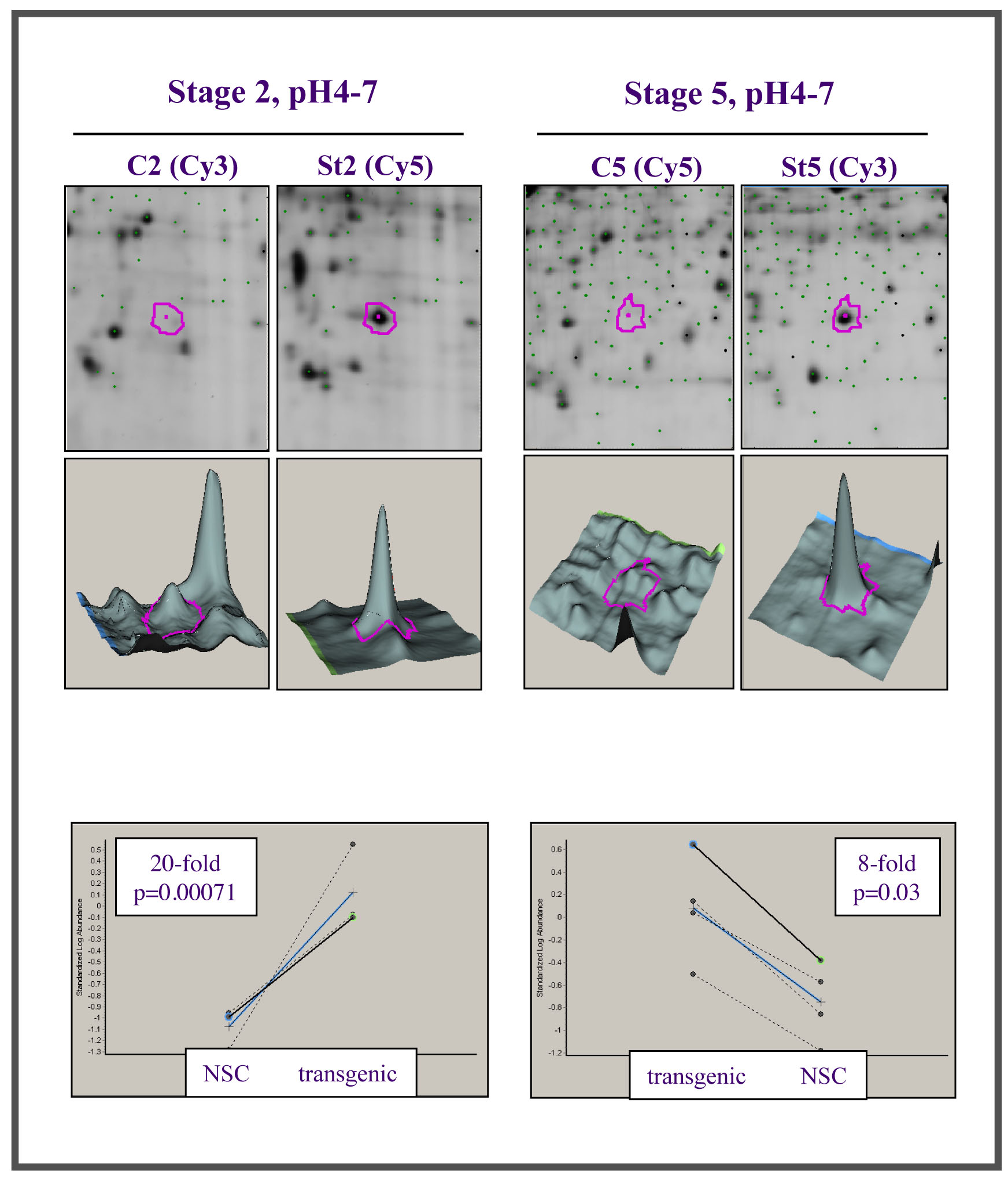
This leads on to another area of active study in the lab. We are exploring the link between chronic inflammation, oxidative stress and cancer. Using a proteomic approach, we have identified a family of proteins, the chitinase-like proteins (Chils), that become hugely upregulated in chronically inflamed tissues (Qureshi et al (2011) Genes and Cancer 2:74-87). We are investigating the role and function of these proteins in chronic inflammation and cancer.
From: Qureshi et al (2011) Genes and Cancer 2:74-87, the data are from a 2D-DiGE study comparing chronically inflamed transgenic tissues (St2 and St5) with controls (C2 and C5 or NSC), to reveal the upregulation (seen as a protein spot (2D) or peak in 3 dimensions, above) of Chil3 and Chil4 (also known as YM1 and YM2) and graphically represented below.