Dr Chris McInerny
Dr Chris McInerny
The role of cell and life cycle specific transcription in fission yeast.
Chris's area of research focuses on the role of gene expression in controlling progression through cell division cycle, using the fission yeast Schizosaccharomyces pombe as a model organism. He is particularly interested in the transcription mechanisms that control the start of S-phase, and the end of the cell cycle at the M-G1 interval.
The fission yeast cell division cycle
Fission yeast has a typical eukaryotic cell cycle with discrete and consecutive phases: G1, S, G2 and M. S-phase is DNA synthesis, M phase is mitosis, and G1 and G2 are 'gap' phases between these two.
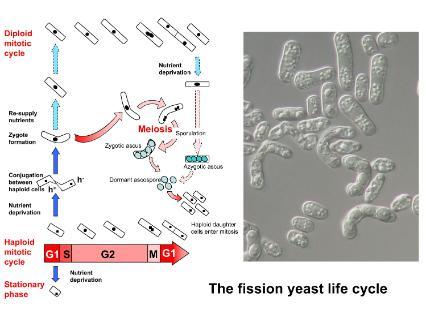
Fission yeast has proved to be an excellent model organism for studying the eukaryotic cell cycle, as many genes and mechanisms first identified in this organism are present throughout the animal kingdom.
Cell cycle transcription
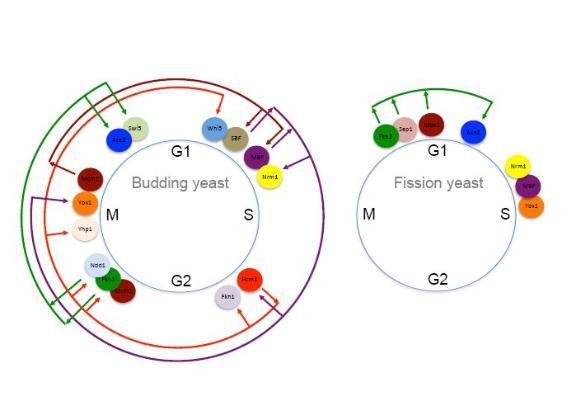
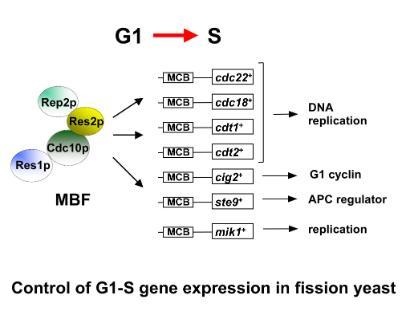
At the beginning of S-phase a group of at least seven genes, including cdc22+, cdc18+, mik1+, cig2+, cdt1+, and cdt2+ are expressed whose products are required, both directly and indirectly, for replicating DNA. The co-ordinate transcription of these genes is controlled by a transcription factor complex binding to a DNA sequence motif present in the promoters of all the genes.
At the start of S-phase the promoter motifs, called MCB (Mlu I cell cycle boxes), are bound by a transcription factor complex called MBF (MCB binding factor; also sometimes called DSC1) containing a number of proteins, including Cdc10p, Res1p, Res2p and Rep2p. Chris is interested in understanding how MBF and MCBs control this cell cycle specific transcription.
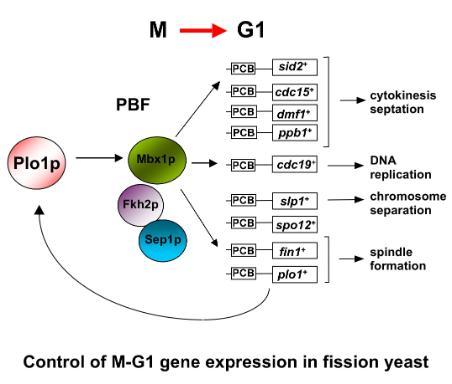
More recently, Chris's group identified another group of genes that is expressed at the M-G1 interval during the fission yeast mitotic cell cycle. This group contains cdc15+, sid2+, ppb1+ and plol+, all of which have functions late in the cell cycle including cytokinesis and septation. These genes contain a motif in their promoters called a PCB (pombe cell cycle box), which binds to transcription factor complex named PBF (PCB binding factor). Chris's group has identified at least three components of PBF: a MADS box protein Mbx1p, and two forkhead like transcription factors, Fkh2p and Sep1p. Strikingly, PBF activity and M-G1 transcription are controlled by the polo-kinase Plo1p through it binding directly to and phosphorylating Mbx1p, so plo1+ controls its own expression in a positive feedback loop. This observation is particularly exciting because polo kinases and MADS box proteins are present in all eukaryotes, including humans, and this suggests a new function for these conserved and important proteins. Chris's group is currently working to further understand how Plo1p regulates this group of genes. Cell cycle and cancer.
Cell cycle transcription in cancer biology
An important part of understanding the basic biology of cancer is to determine how normal cell division is regulated. Yeast is an excellent model system in which to study the cell division cycle as many genes essential for its control, originally identified in this organism, are present throughout nature.
It is now known that networks of cell cycle transcription occurs in all eukaryotic organisms, encompassing the whole cell cycle. In some cases the transcription factors are mutated in various forms of cancer. Thus, the ultimate goal of Chris's research is to analyse the role of transcription in cancer biology [1].
doi: 10.12688/f1000research.8111.1. eCollection 2016.