Prof Barbara Mable
Prof Barbara Mable
 Professor of Evolutionary Genetics
Professor of Evolutionary Genetics
Institute of Biodiversity, Animal Health & Comparative Medicine
College of Medicine, Veterinary & Life Sciences
Graham Kerr Building
University of Glasgow
Glasgow, G12 8QQ
Tel.: +44 (0)141 330 3532
Email: Barbara.Mable@glasgow.ac.uk
Why Study Polyploids?
Why Study Polyploids?
Polyploidy, the presence of more than two copies of each chromosome in the cells of an individual organism, has undoubtedly been a significant force driving genetic diversity and speciation in the evolutionary history of plants, and recent evidence suggests that it is more important in animal evolution than was previously thought. In fact, evidence from genome sequencing projects suggests that even in organisms with small genomes (the yeast Saccharomyces cerevisiae and the plant Arabidopsis thaliana) there is evidence of ancient genome duplication events. This has lead to a dramatic increase in study of the potential fate of duplicate genes, and has brought polyploidy from a somewhat peripheral plant-only perspective to the main stream of comparative genomics, bioinformatics, and evolutionary research. Recent technological advances have greatly enhanced the potential to elucidate both the origins and consequences of polyploidy (and gene duplication in general) in a wide array of organisms.
I have studied the evolution of polyploidy in animals (frogs in the genera Hyla and Neobatrachus and salamanders in the genus Ambystoma), plants (flowering plants in the genus Arabidopsis), and fungi (yeast in the genus Saccharomyces) (Figure 1). My research has combined classical Mendelian genetics with more advanced technologies, including: comparative sequence analysis (PCR, cloning, sequencing), rtPCR-based gene expression studies, genomic-scale expression studies using microarrays, genome content analysis using flow cytometry, cytogenetics using image analysis, and inheritance and diversity studies using microsatellite DNA. We are also increasingly using data obtained from next generation sequencing approaches, including 454 tagged amplicon sequencing, Illumina-based bulked segregant analysis, and Restriction Associated DNA (RAD) sequencing. We are also increasingly using data obtained from next generation sequencing approaches, including 454 tagged amplicon sequencing, Illumina-based bulked segregant analysis, MiSeq genotyping of large gene fragments, Restriction Associated DNA (RAD) sequencing and low coverage whole genome resequencing.
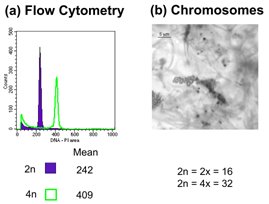
Figure 1. Techniques used to assess ploidy level. (a) Flow cytometry; (b) chromosome counts (Based on Dart, Kron & Mable, 2004. Can J. Bot.).
Previous Research
Previous Research
1) Plant Mating Systems
i) Evolutionary dynamics of genes controlling self-incompatibility in natural populations
Sporophytic self-incompatibility (SSI), which is controlled by genes that belong to large gene families, has been extensively studied in cultivated plants in the genus Brassica. In a continuing collaborative project with the research groups of Deborah Charlesworth (University of Edinburgh, Scotland) and Mikkel Schierup (University of Aarhus, Denmark) we characterized the system in naturally occurring populations of Arabidopsis lyrata, which is related both to Brassica and to the self-compatible genetic model, Arabidopsis thaliana and have extended it to other species in the Brassicaceae (in collaboration with Xavier Vekemans, University of Lille; Steve Ansell, Natural History Museum London; Detlef Weigel, Max Planck Institute for Developmental Biology, Tübingen; Marcus Koch, University of Heidelberg). We identified a large number of S-alleles from the stigma-expressed SRK allele (the female S-gene) as well as additional paralogous loci that do not appear to be directly involved in the SI reaction (Figure 1). Some of the intriguing aspects of self-incompatibility systems are i) the spectacular polymorphisms maintained in populations over long evolutionary times and ii) the complex dominance systems that appear to operate in sporophytic systems. We were particularly interested in using this gene family as a model for understanding the evolutionary dynamics of genes under balancing selection (negative frequency dependent selection). One challenge is in genotyping highly polymorphic gene families; we have attempted various approaches, including working with the NERC Biomolecular Analysis Facility in Liverpool to develop other barcode-based approaches (using MiSeq) for SI genotyping that are less error prone than originally attempted 454 sequencing.
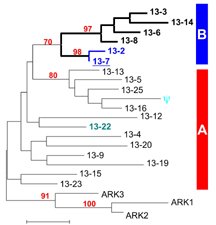
Figure 1. Gene tree showing relationships among a subset of alleles at the self-incompatibility locus of A. lyrata. Sequences can be divided roughly into two major groups (A and B). (Based on Prigoda, Nassuth & Mable, 2005. Mol. Biol. Evol.).
One of my particular interests has been investigation of the consequences of polyploidy on mating system variation and the gene families involved in these types of recognition processes. In some systems of SI, polyploidy is often accompanied by increased levels of self-compatibility but this pattern does not seem to be very strong in SSI systems (Mable 2004. New Phyt.). We have found both strongly SI (Europe) and completely selfing (Asia) tetraploid populations of A. lyrata (Figure 2). We have also established that similar dominance of S-alleles is found in SI tetraploids, compared to that in SI diploids (Mable et al. 2004 Heredity; Mable et al. 2018 Frontiers in Ecology & Evolution). This work involves a combination of establishment of ploidy level using flow cytometry (Dart et al. 2004 Canadian Journal of Botany), controlled hybridisation experiments in the greenhouse to establish the genetics of SSI and extensive molecular characterization (through PCR, cloning and Sanger sequencing or deep sequencing approaches) of the number of S-alleles present in natural populations, their relative age, and divergence levels compared to alleles at loci that are members of the same gene family but that are not involved in SI, and differences in expression and dominance of alleles of different types.
In collaboration with Marte Jørgensen, Karin Lagesen and Anne Brysting from the University of Oslo, we used 454 tagged amplicon sequencing for genotyping the complex gene family associated with the female component of SI (S-related kinase, SRK) in diploids and polyploids occurring in a hybrid zone between A. lyrata and Arabidopsis arenosa that has been studied extensively by Marcus Koch and his group (Jørgensen et al. 2013 Plant Ecology & Diversity). We used this gene family to investigate the consequences of hybridisation and polyploidy on genetic diversity of gene families under balancing selection, using a combination of 454 sequencing and more classical methods (Mable et al. 2018 Frontiers in Ecology and Evolution[BM1] ). We also worked with the NERC Biomolecular Analysis Facility in Liverpool to develop other barcode-based approaches (using MiSeq) for SI genotyping that are less error prone than 454 sequencing.

Figure 2. Morphological differences between diploid and tetraploid A. lyrata. (a) Diploid plant (2x=16) from Iceland; (b) tetraploid plant (4x=32) from Austria; (c) comparison between diploid (upper) and tetraploid (lower) flower sizes; (d) comparison between diploid (upper) and tetraploid (lower) leaf rosettes.
ii) Break-down of SI in natural diploid populations
Despite reports that A. lyrata is an exclusively self-compatible species, we unexpectedly discovered that this is not true for all populations. We have identified diploid and tetraploid populations that appear to be completely self-compatible, whereas others consist of completely self-incompatible individuals or a mixture of self-compatible and self-incompatible individuals. We have established that loss of SI has resulted in a substantial loss of genetic diversity and that there has been a shift to inbreeding in some populations (Mable et al. 2005 Evolution; Mable & Adam 2007 Molecular Ecology).
AD-sequencing to characterise genome-wide diversity in relation to mating system (Buckley et al. 2018 BMC Genomics) and 6) investigating whether the shift to inbreeding has compromised short-term adaptive potential (Buckley et al. 2019 BiRxiv). These populations are found on threatened habitats (sand dunes, cliff edges, and limestone pavements: Figure 3) and results from this work may have conservation implications for the effects of habitat fragmentation on plant mating systems. This work is being continued in collaboration with one of the original postdocs involved with the project, Marc Stift, from the University of Konstanz. This work involved four postdocs: Hong-Guang Zha, Marc Stift, Annabelle Haudry and James Buckley; three PhD students: Peter Hoebe, Andy Tedder, and Paul Foxe (York University, Toronto); and two main collaborators: Dr. Stephen Wright (University of Toronto) and Steve Ansell (Natural History Museum, London). We also worked with Sang-Tae Kim, Jürg Hagmann and Detlef Weigel in Tübingen to investigate the genetic basis for loss of SI using an Illumina-sequencing-based bulked segregant approach (Mable et al. 2018 Heredity).

Figure 3. A. lyrata habitats. (a) Rocky ridge, Ise of Skye, Scotland; (b) Cliff edge, Tobermory, Ontario; (c) Alvar, Tobermory, Ontario; (d) Sand dune, Rondeau Provincial Park, Ontario.
iii) Polyploidy and Self-incompatibility
Some of the intriguing aspects of self-incompatibility systems are i) the spectacular polymorphisms maintained in populations over long evolutionary times and ii) the complex dominance systems that appear to operate in sporophytic systems. These phenomenon have not been described for polyploids. In some systems of SI, polyploidy is often accompanied by increased levels of self-compatibility but this pattern does not seem to be very strong in SSI systems (Mable 2004. New Phyt.). We have found both strongly SI (Europe) and completely selfing (Asia) tetraploid populations of A. lyrata. We have already established that similar dominance of S-alleles is found in SI tetraploids, compared to that in SI diploids (Mable et al. 2004 Heredity) but we would like to look at this in more detail, using both diallele crosses and rtPCR to examine allelic expression patterns in diploids and tetraploids that share the same alleles in order to determine how increased copy number influences relative compatibility. Ploidy of individuals has been determined using flow cytometry and confirmed by chromosome counts for representatives from each population that we have sampled.
In collaboration with Marte Jørgensen, Karin Lagesen and Anne Brysting from the University of Oslo, we have been using 454 tagged amplicon sequencing for genotyping the complex gene family associated with SI in diploids and polyploids occurring in a hybrid zone between A. lyrata and Arabidopsis arenosa that has been studied extensively by Marcus Koch (Jørgensen et al. 2013 Plant Ecology & Diversity). I have also been working with the NERC Biomolecular Analysis Facility in Liverpool to develop other barcode-based approaches (using MiSeq) for SI genotyping that are less error prone than 454 sequencing.
 Figure 4. Morphological differences between diploid and tetraploid A. lyrata. (a) Diploid plant (2x=16) from Iceland; (b) tetraploid plant (4x=32) from Austria; (c) comparison between diploid (upper) and tetraploid (lower) flower sizes; (d) comparison between diploid (upper) and tetraploid (lower) leaf rosettes.
Figure 4. Morphological differences between diploid and tetraploid A. lyrata. (a) Diploid plant (2x=16) from Iceland; (b) tetraploid plant (4x=32) from Austria; (c) comparison between diploid (upper) and tetraploid (lower) flower sizes; (d) comparison between diploid (upper) and tetraploid (lower) leaf rosettes.
2) Resistance gene evolution in plants
At least some types of resistance genes in plants evolve under the same types of selection pressures as self-incompatibility genes (i.e. balancing selection), when the particular host-pathogen interaction involves negative frequency dependent selection. We used A. lyrata as a model to investigate how mating system and polyploidy affect such pathogen response systems, focusing predominantly on its interactions with oomycete pathogens known to be problematic for cultivated species in the Brassicaceae. In collaboration with Eric Holub, at the University of Warwick, we investigated responses to pathogens in the genus Albugo (causing white blister rust, Figure 4), in relation to ploidy and mating system in the host and the consequences of these host characteristics for diversity at candidate pathogen resistance genes. We also used RAD sequencing to enable comparison of genome-wide patterns of variation in neutral genes with those that might be involved in adaptation (Buckley et al. 2016) The project involved three postdoctoral researchers (Volkan Cevik and Joana Vicente, Warwick; James Buckley, Glasgow), and three research technicians (Aileen Adam, Elizabeth Kilbride, Ryan Carter).
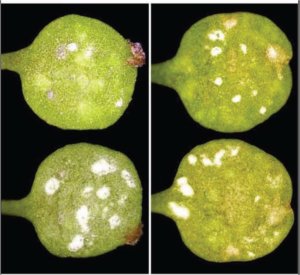
Figure 4. A. lyrata plants infected with Albugo candida
3) Immune gene variation in polyploids
One consequence of polyploidy that has received remarkably little attention is how whole genome duplication affects the ability to respond to pathogens. Although theories suggest that the greater genetic diversity provided by having twice as many gene copies should increase potential to respond to a wider array of pathogens, this is contradicted by theories of immune gene variation, which predict that having too many copies of immune genes could actually be detrimental. Tetraploid frogs in the genus Xenopus (African clawed frogs; Figure 5) appear not to retain all four copies of genes at the Major Histocompatibility complex but this has only been investigated in lab strains, where they appear to retain three copies. In collaboration with Richard Tinsley and Mark Viney from the University of Bristol, we investigated variation at the MHC and neutral genes in feral populations of Xenopus that had been living in Wales for at least 40 years. Although the population was extirpated by extreme weather conditions in 2010, we were able to confirm that they originated from laboratory strains originally collected in the Western Cape region of South Africa (Mable et al. 2015 Immunogenetics). Despite origination from a lab population, surprisingly high levels of diversity were maintained not only at the MHC and other immune genes, but also at neutral loci. In addition, there was variation among individuals in the number of MHC gene copies retained (between 1 and 3). This emphasizes the increased genetic flexibility possible in polyploids, even in the face of a severe bottleneck of inbred lab strains.
Figure 5. Xenopus laevis.

