Recent highlights in understanding heterochromatin barrier activity
We have found that the heterochromatin barrier activity of the HS4 element is mediated by binding sites for the proteins USF1/USF2 and VEZF1 (CTCF binding is not required), which direct different mechanisms to counter the propagation of chromatin silencing. The USF transcription factors act to recruit several histone modifying enzymes that result in the acetylation of H3, H4 and H2A.Z and the methylation of H3K4 and H4R3. These so-called active histone modifications are proposed to collectively act as a chain terminator to heterochromatin assembly by interfering with the propagation of repressive histone modifications.
H2B Ubiquitination is Required for Chromatin Boundary Integrity
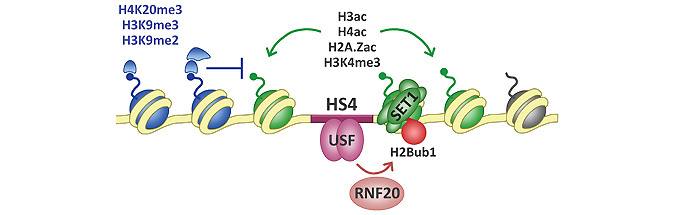 The HS4 insulator element marks the 3′ boundary of a heterochromatin region located upstream of the chicken β-globin gene cluster. We found that USF1 recruits the E3 ligase RNF20/BRE1A to mediate H2B mono-ubiquitination (H2Bub1) at the HS4 insulator (Ma et al., 2011. PLoS Genetics, 7(7) e1002175 ). Knockdown experiments show that RNF20 is required for H2Bub1 and processive H3K4 methylation. Depletion of RNF20 results in a collapse of the active histone modification signature at the HS4 chromatin boundary, where H2Bub1, H3K4 methylation, and hyperacetylation of H3, H4, and H2A.Z are rapidly lost. A remarkably similar set of events occurs at the HSA/HSB regulatory elements of the neighbouring FOLR1 gene, which mark the 5′ boundary of the same heterochromatin region.
The HS4 insulator element marks the 3′ boundary of a heterochromatin region located upstream of the chicken β-globin gene cluster. We found that USF1 recruits the E3 ligase RNF20/BRE1A to mediate H2B mono-ubiquitination (H2Bub1) at the HS4 insulator (Ma et al., 2011. PLoS Genetics, 7(7) e1002175 ). Knockdown experiments show that RNF20 is required for H2Bub1 and processive H3K4 methylation. Depletion of RNF20 results in a collapse of the active histone modification signature at the HS4 chromatin boundary, where H2Bub1, H3K4 methylation, and hyperacetylation of H3, H4, and H2A.Z are rapidly lost. A remarkably similar set of events occurs at the HSA/HSB regulatory elements of the neighbouring FOLR1 gene, which mark the 5′ boundary of the same heterochromatin region.
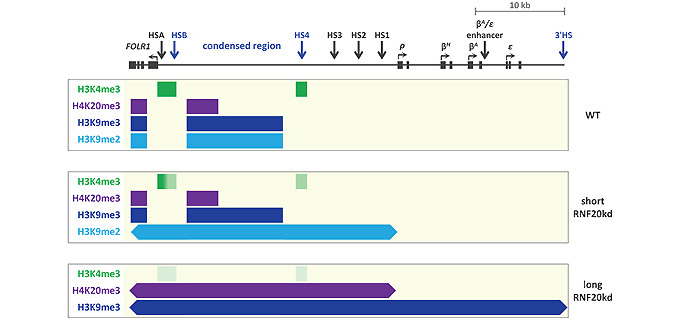
Persistent H2B ubiquitination at the HSA/HSB and HS4 elements is required to maintain chromatin boundary integrity. Depletion of H2B ubiquitination results in the sequential spreading of the repressive chromatin marks H3K9me2, H3K9me3, and H4K20me3 over the entire 50 kb FOLR1 and β-globin region. This heterochromatin spreading results in the progressive silencing of FOLR1 gene transcription. These findings show that the HSA/HSB and HS4 boundary elements direct a cascade of active histone modifications that defend the FOLR1 and β-globin gene loci from the pervasive encroachment of an adjacent heterochromatin domain. We are now studying a number of mammalian gene loci that employ H2Bub1-dependent boundaries to prevent heterochromatin spreading.
Insulator sequences induce early DNA replication timing
 The temporal programme of DNA replication during S-phase generally mirrors gene expression programmes in vertebrates. It is considered that the same processes that mediate the organisation chromosomal domains that favour gene expression may also control DNA replication timing, but these mechanisms are poorly understood. We have worked with the group of Marie-Noëlle Prioleau, Paris to address whether insulator elements contribute to replication timing.
The temporal programme of DNA replication during S-phase generally mirrors gene expression programmes in vertebrates. It is considered that the same processes that mediate the organisation chromosomal domains that favour gene expression may also control DNA replication timing, but these mechanisms are poorly understood. We have worked with the group of Marie-Noëlle Prioleau, Paris to address whether insulator elements contribute to replication timing.
We found that the HS4 insulator element has the capacity to shift a late replicating origin to earlier replication. This timing activity was mapped to HS4’s USF binding site, which itself is sufficient to drive earlier replication. The USF activity is associated with the recruitment of the active histone modifications H3ac, H3K4me3 and H2A.Zac. The timing shift can be enhanced further by the presence of a transcriptionally active gene. These results show that combinations of elements that enrich for active histone modifications, and perhaps favour nuclear localisation away from the periphery, favour the accessibility to S-phase kinases required for early replication.
Control of DNA methylation by VEZF1
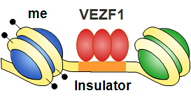 While USF-mediated histone modifications are an essential component of HS4’s barrier activity, they are not sufficient. We recently found that the transcription factor VEZF1 interacts with three sites of the HS4 insulator that are all required for its barrier activity (Dickson et al., 2010. PLoS Genetics 6 (1):e1000804 ). Neither of these sites is required for the recruitment of active histone modifications. Rather, the VEZF1 sites are required to prevent DNA methylation. VEZF1 is a candidate CpG island protection factor as the G-rich sequences bound by VEZF1 are commonly found at CpG island promoters. For example, we found that VEZF1 binding elements are sufficient to mediate demethylation and protection of the APRT CpG island promoter from DNA methylation (Dickson et al., 2010). These findings are an important development for understanding chromatin boundary formation and CpG island regulation in vertebrates. We are currently studying VEZF1 interacting proteins and determining their roles in the control of DNA methylation.
While USF-mediated histone modifications are an essential component of HS4’s barrier activity, they are not sufficient. We recently found that the transcription factor VEZF1 interacts with three sites of the HS4 insulator that are all required for its barrier activity (Dickson et al., 2010. PLoS Genetics 6 (1):e1000804 ). Neither of these sites is required for the recruitment of active histone modifications. Rather, the VEZF1 sites are required to prevent DNA methylation. VEZF1 is a candidate CpG island protection factor as the G-rich sequences bound by VEZF1 are commonly found at CpG island promoters. For example, we found that VEZF1 binding elements are sufficient to mediate demethylation and protection of the APRT CpG island promoter from DNA methylation (Dickson et al., 2010). These findings are an important development for understanding chromatin boundary formation and CpG island regulation in vertebrates. We are currently studying VEZF1 interacting proteins and determining their roles in the control of DNA methylation.

