Dr Drew Thomson
- Lecturer (School of Chemistry)
telephone:
01413308284
email:
Drew.Thomson@glasgow.ac.uk
Room A3-15, Joesph Black Building, School of Chemistry, G12 8QQ
Biography
Drew Thomson
I started my independent career as a Lecturer in Chemical Biology at Glasgow in 2016. Before this I studied for an MChem degree at University of Edinburgh, where I then stayed for a PhD in supramolecular organic chemistry in the group of Prof David Leigh. After a short postdoc in protein biophysics (also in Edinburgh) in the group of Dr David Dryden, I moved to Bristol, as a postdoc and later research fellow, in the lab of Prof Dek Woolfson, working on peptide design and synthesis. The Thomson group now works on combining all these disparate influences to understand and design peptides in chemical biology.
Outside of research, I am enthusiastic about widening participation in chemistry. I am now the Athena SWAN lead for Chemistry at University of Glasgow, and am keen to learn about, discuss, promote and help with any activities around EDI in science.
Publications
Google Scholar link to Drew's publications
Reviewing
I review regularly for RSC and ACS journals, and am always keen to review manuscripts at the chemistry/biology interface, and especially in the peptide chemistry and chemical biology spheres. I am a member of the EPSRC College, and review grant applications from EPSRC, BBSRC and MRC, as well as the RSC and Leverhulme Trust.
Work with us
All of our funded studentship positions are filled at the moment. If you have, or are applying for, your own PhD or postdoctoral fellowship funding and are looking for a host group please do get in contact. We are happy to support applications for individual funding.
Research interests
Overview
Research in our lab is aimed at designing peptides with defined 3D conformations. We use these peptides to ask questions about biological systems, and to mimic behaviour seen in biology. By designing these molecules from first principles, we learn about the fundamental factors that govern the relationship between atomic connectivity and three-dimensional structure. Work in this area is necessarily multidisciplinary: our lab uses a mix of bioinformatics, computational modelling, peptide and organic synthesis, and biophysical characterisation. Our lab has all the resources needed for the synthesis, purification, and analysis of peptides, as well as resources and expertise in computational design and modelling. We are always interested to discuss potential collaborations! If you have a research question that could benefit peptide synthesis or design then please do get in contact.
Research Projects
Some of the projects we are currently researching in the Thomson lab:
Non-native Chemical Ligation
We have recently become interested in mimicking aspects of protein structure using the residual chemical functionality from a ligation reaction. This allows us to construct a beta turn mimic at the same time as carrying out a ligation reaction. In doing so, we generate a peptide which contains a non-natural unit, but which replicates the structural role of the residues that it has replaced. We are now expanding this method beyond beta turns to other linking units, and are investigating using these turn mimics as a way of adding extra chemical functionality to a peptide/protein mimic.
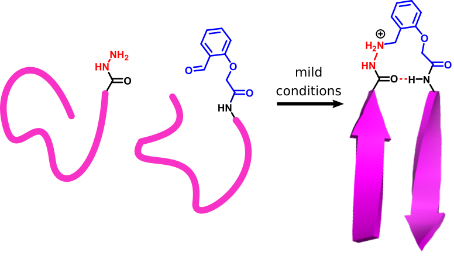
Cyclic Peptide Conformational Design
Cyclic peptides are an interesting sub-category of peptide because they are often exhibit fewer conformations than their linear counterparts. For this reason they have a lot of potential as bioactive molecules for research in Chemical Biology. However, designing a peptide sequence so that a cyclic peptide adopts a particular conformation is still a challenging task. We are investigating methods to address this challenge, and are testing these methods by developing cyclic peptides that mimic known binding loops in natural systems.
Computational Protein Design
We are generally interested in computational methods that can simplify the challenge of understanding the otherwise often bewildering behaviour of peptides and proteins. In particular, we are particularly interested in protein systems such as beta hairpins, collagen-like peptide, alpha helical coiled coils and others, where the geometry of the peptide can be described using comparatively few parameters. This allows us to use computer models to design and test new structural variants. We are contributors to the "Isambard" Python module for peptide/protein design and analysis.
Hybrid Peptide Biomaterials
Nature uses peptides/proteins for a range of structural roles, and there is a great deal yet to be learned about how these evolved materials. As part of a project with the LifETIME CDT, we are working with the group of Dr Bernhard Schmidt to investigate new peptide/polymer hybrid materials as scaffolds for cell engineering.
Research groups
Grants
We have funding through EPSRC grants and via the Leverhulme Trust.
Supervision
PhD Students
Selma Crecente Garcia
PhD Student 2017-2021
Selma works on the design of beta turn mimics and peptides that exhibit conformational switching behaviour. Selma has skills in peptide synthesis, peptide/protein NMR, and peptide biophysics.
Bethany Atkinson
PhD Student 2018-2022
Bethany works on the conformational design of cyclic peptides, as well as peptide ligation methods. Bethany is an expert in enhanced sampling molecular dynamics methods, as well as peptide synthesis and biophysical analysis.
Pernille Christensen
PhD Student 2020 -2023
Pernille is working on methods for generating mimics of helical proteins through chemical ligation reactions. This involves a mix of organic synthesis, peptide chemistry, and protein biophysics methods.
Robbie Stirling
PhD Student 2021-2024
Robbie will be joining in October 2021 for an EPSRC funded studentship that will explore adding different chemical functions to our chemical ligation methods to make new probe molecules for chemical biology.
Cameron McAnespie
PhD Student 2021-2024
Cameron will be joining in October 2021 through the LifETIME CDT, and will be working on hybrid peptide/polymer materials for tissue engineering applications.
Danielle Liebnitz
PhD Student 2021-2024
Danielle will join us for a Leverhulme Trust funded studentship starting Oct 2021, joint with the group of Prof Ross Forgan. The project will involve making new and interesting chiral peptide/inorganic hybrid structures.
Postdoctoral Researchers
Beckie Clarke
PDRA 2019-2023
Beckie works jointly with the group of Prof Andy Sutherland on the synthesis of fluorescent amino acids and their incorporation into designed peptides/proteins. These will form the basis of new chiroptical sensing methods. This is part of an EPSRC funded project led by Prof Malcolm Kadodwala
Teaching
Level 1: Organic Chemistry 2
Level 3: Organic Synthesis 1
Chemical Biology PGT: Joint Course Coordinator and Bioconjugation Lecture course.



